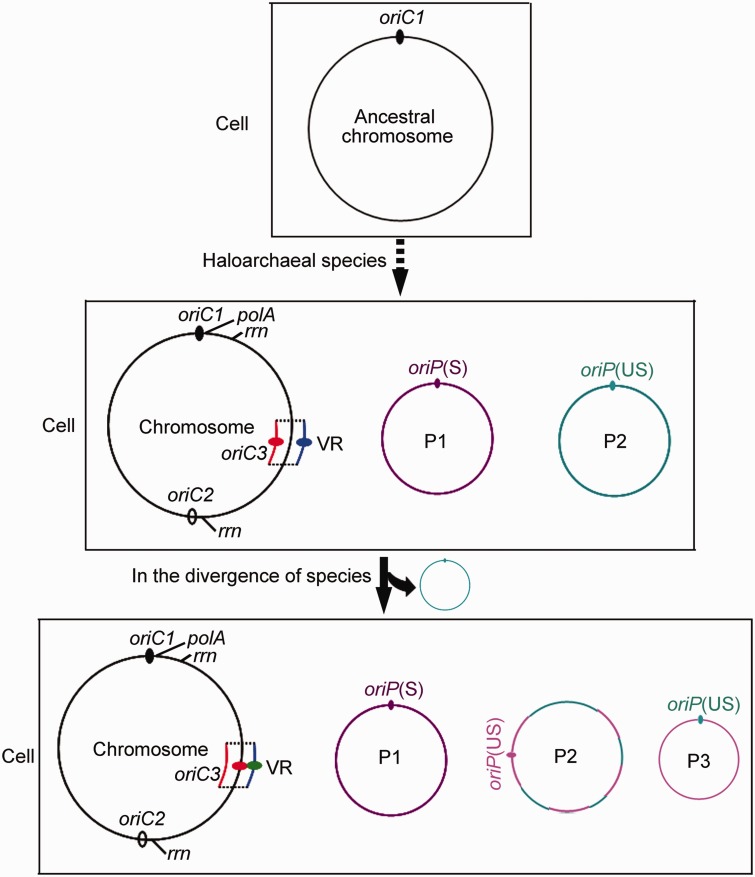
Fig. 6.—
Model of the association between the dynamics of replication origins and genome evolution. The haloarchaeal multireplicon genome evolved in multiple steps (indicated with the dotted arrow) from the oriC1-dependent archaeal ancestral chromosome (up). For example, novel replication origins (oriC2 and oriC3) were independently integrated into the chromosome, and minireplicons were constructed in the branch leading to specific haloarchaeal species, whereas oriC1 was conserved via its effects on surrounding genes (middle). Both the replication origins and the genome architecture (particularly the extrachromosomal replicons) varied frequently in the divergence of species (down). The oriC2 origin was conserved in a similar manner as oriC1 via its effects on surrounding genes, whereas oriC3 was continually being acquired, lost and disrupted in the chromosomal variable regions (in different colors). The fate of extrachromosomal replicons was determined by the hierarchy in the plasmid stabilization of their bearing replication origins. Conversely, unstable extrachromosomal replicons forced variation of their bearing replication origins. For example, the plasmid P1 (in purple) was conserved, as was the stability of the ARS plasmid with its bearing origin oriP(S). In contrast, the plasmid P2 (in dark green) was easily lost because the oriP(US)-based ARS plasmids were unstable. However, parts of the P2 contents might be maintained via the reconstruction of a novel P2 plasmid together with novel genes in the surrounding environments (in mosaic of dark green and pink). In addition, the oriP(US) can be used to construct the novel plasmid (P3) with its surrounding genes (in pink).