Abstract
Promoter hypermethylation is associated with loss of expression of tumor suppressor genes in cancer. Currently, several genome-wide technologies are available and have been utilized to examine the extent of DNA methylation in discovery-based studies involving several physiological and disease states. Although early in the process, aberrant DNA methylation is gaining strength in the fields of cancer risk assessment, diagnosis, and therapy monitoring in different cancer types.
There is a need to improve existing methods for early diagnosis of prostate cancer (CaP) and to identify men at risk for developing aggressive disease. Because of the ubiquity of DNA methylation changes and the ability to detect methylated DNA in several body fluids (blood, urine), this specifically altered DNA may serve, on the one hand, as a possible new screening marker for CaP and, on the other hand, as a tool for therapy monitoring in patients having had neoplastic disease of the prostate. As many CaP patients present with advanced disease or some of the men with non-specific elevation of PSA without CaP, early detection with high-specificity and sensitivity to be one of the most important approaches to reduce mortality and unwanted tension of the men with high PSA. Therefore, an effective screening test would have substantial clinical benefits. Furthermore, methylation markers of risk of progression of disease in patients having CaP permits immediate commencement of specific treatment regimens and probably longer survival and better quality of life. This review illustrates the current benefits and limitations of potentially useful CaP methylation markers that have considerable existing data and touches upon other future markers as well as the field of methylation in CaP.
Keywords: Prostate Cancer, DNA methylation, Urine, Serum, Screening, Therapy monitoring, Review
The successful completion of the Human Genome Project (HGP) has resulted in the annotation of around 30,000 genes. The major key question facing the scientific and clinical communities has been how to use this information for the benefit of cancer patients such as early detection, prognosis, therapeutic response and monitoring of disease. Hypermethylation or hypomethylation of the promoter region (one of the epigenetic alteration) inactivate or activate certain genes that are related to cancer. Different approaches to study promoter region methylation have been developed in recent years. Information from HGP facilitates these approaches. This Review describes the different ways that methylation of the promoter region of different key cancer gene/genes that have led to the identification of novel biomarkers that have affected, or that have the potential to affect, the clinical management of human prostate cancer. A survey of biomarkers identified by other approaches is provided elsewhere (1).
The Clinical Problem of Prostate cancer
In the US, prostate cancer is the most common cancer in men and a major cause of cancer death (2). Identifying biomarkers and new molecular targets has the potential to greatly improve the management of prostate cancer, as the current procedures used in the initial diagnosis, including PSA detection, digital rectal examination, trans-rectal ultrasonography and histopathological examination of biopsy specimens, have limited sensitivity and specificity. In particular, clinical objectives such as improved accuracy of detection and diagnosis of prostate cancer, the identification of markers of aggressive disease, and the discovery of new targets for cancer therapy could be facilitated by better biomarkers. Once the cancer has been diagnosed, there are also significant problems with the management of this disease; for example, the natural history of development of prostate cancer is highly variable. A significant proportion of cancer detected by PSA screening might never become life threatening in the absence of treatment, and over-treatment of this cancer group represents a major concern leading to unwanted suffering of the patients and increases substantial morbidity (3). Alternatively, more conservative approaches might leave aggressive cancers untreated that also increases prostate cancer related death. Novel biomarkers that allow patients with indolent cancer to be spared inappropriate treatment, and that facilitate the targeting of radical therapies towards aggressive disease are, therefore, urgently required.
As primary prostate cancers are dependent on androgens for their growth, patients who have advanced disease are often treated with androgen withdrawal therapies. Success is variable but the treatments eventually fail, leading to the development of castration-resistant prostate cancer (CRPC) and usually to death. A better understanding of the molecular pathways that support the androgen-dependent growth of prostate cancer and of the mechanism of development of CRPC are therefore required, so that improved strategies for managing advanced disease can be developed.
There is a tremendous need for non-invasive detection strategies for prostate cancer
Prostate cancer is the most common non-dermatologic malignancy in men and the second leading cause of cancer-related death in the United States, with a projected incidence of 186,320 new cases and 28,660 deaths due to carcinoma of the prostate in 2008 (NCI webpage: http://www.cancer.gov/cancertopics/types/prostate). Prostate cancer is curable if detected early, while still localized within the capsule (4).Radical retro-pubic prostatectomy (RRP) is an effective treatment for clinically localized prostate cancer. Despite the earlier detection of prostate cancer, recurrent prostate cancer remains an important problem in the long-term in approximately 40% of patients who undergo RRP, with most of the relapses (95%) in the first 5 years (5). Diagnosis and management of recurrent prostate cancer are confounded by the lack of symptoms and the lack of cancer-specific diagnostic techniques to be used during early stages of disease. To date, curative therapeutic options for the majority of patients depend on early detection, including digital rectal examination and measurement of prostate-specific antigen (PSA) in the serum. PSA is regarded as one of the best conventional serum tumor markers; however, determination ofPSA levels alone is neither sensitive nor specific enough for a definite diagnosis of prostate cancer (6, 7). Most men with either an abnormal finding on digital rectal examination or elevated PSA require trans-rectal biopsies with ultrasound guidance, and up to one third of these men are found to be free of disease. Moreover, by this approach, false-negative biopsies still occur in approximately 25% of patients. Novel approaches for the definitive detection and control of recurrent prostate cancer are urgently needed.
In addition to an impact on early detection, the discovery of better molecular markers of prostate cancer could have a significant impact on our management of the disease by facilitating a rapid determination of tumor responses to novel therapies. Molecular markers can potentially be used to diagnose early cancers and perhaps even pre-invasive lesions of the prostate. Recognition of pre-invasive lesions of prostate carcinoma could ultimately enable clinicians to treat a neoplasm before it invades tissues. Just as screening for pre-invasive neoplasms in the cervix and breast have saved the lives of many women, so too may the detection of precursor lesions in the prostate save the lives that would otherwise be lost to invasive prostate cancer. Therefore, once we identify molecular markers selectively altered in early recurrent prostate carcinoma, we will examine the alterations of these markers in the potential precursors to invasive prostate carcinoma to determine when their aberrant alteration occurs in prostate neoplasia.
DNA Methylation abnormalities are widespread in cancer and DNA methylation abnormalities have been detected in patients with early-stage cancer
Changes in the status of DNA methylation, known as epigenetic alterations, are one of the most common molecular alterations in human neoplasia (8-11) including prostate cancer (12-16). Cytosine methylation occurs after DNA synthesis by enzymatic transfer of a methyl group from the methyl donor S-adenosylmethionine to the carbon-5 position of cytosine. Cytosines are methylated in human genome mostly when located 5′ to a guanosine. Regions with a high G: C content is so called CpG islands. It has been increasingly recognized over the past four to five years that the CpG islands of a large number of genes, which are mostly unmethylated in normal tissue, are methylated to varying degrees in human cancers, thus representing tumor specific alterations (17, 18).The presence of abnormally high DNA concentrations in the serum, plasma and urine of patients with various malignant diseases was described several years ago (19-21) and recently specifically reviewed for genitourinary cancer by Paul cairns(22). The discovery that cell-free and cell-bound DNA can be shed into the lumen of the organ of various malignancies has generated great interest. During recent years an increasing number of studies have reported the presence of methylated DNA in bodily fluids of various types of malignancies (23-26) (14,15,16,17) including urine of prostate cancer patients at diagnosis and the absence of methylated DNA in normal control patients (12). DNA methylation alterations are particularly amenable to sensitive detection. In contrast, to detect genetic mutations one must overcome the problem of a large number of possible mutations.
DNA methylation in prostate cancer
The link between methylation at the C5-position of cytosine in CpG sequences and cancer development is well established. Cancer formation is accompanied by dramatic changes in the cellular methylation profile such that global demethylation of the genome occurs in parallel with CpG hypermethylation at specific genes strongly linked to their transcriptional inactivation. A list of genes that are hypermethylated at CpG islands in prostate cancer is shown in Table 1. Several of these genes, including RAR-β2, INK4a, RASSF1a and APC, exhibit tumor suppressor functions whose inactivation associated with hypermethylation of CpG islands in 5" regulatory regions occurs during prostate cancer development (27-33). Inappropriate gene hypermethylation catalyzed by DNA methylases (DNMTs) may represent an early event in cancer development, possibly linked to ageing. Promoter Methylation of GSTpi was absent in normal epithelium and present in 6.4% of proliferative inflammatory atrophy, in 70% of high-grade PIN and in 90% of prostate cancer (34). When methylation at the APC gene was considered together with methylation of GSTpi, the sensitivity for detecting cancer approached 100% (32). Methylation may also be associated with tumor progression. For example, CpG hypermethylation of the cell adhesion gene E-cadherin in breast and prostate is integral to epithelial-to-mesenchymal transition that is believed to play a prominent role in tumor progression (35). Methylation of the oestrogen receptor alpha (ESR1) gene, whose down-regulation has been suggested to play a role in cancer metastasis, has also been documented in prostate cancer (36). Hypomethylation of specific genes is also linked to prostate cancer development. For example, Wang et al (37) have reported hypomethylation of WNT5A, CRIP1, and S100P in cancer but not in normal prostate. Interestingly, CpG methylation status appeared to control binding of MYB to the WNT5A promoter region. Our unpublished data in several cancer types indicate that there are some groups of genes that are methylated in normal but not in cancer. The details mechanism of this group of gene in cancer development needs to be explored.
Table 1.
Genes showing frequent hypermethylation in human prostate cancer
| Gene | HR | CCC | RAD | ST | CBM | IR | CTSG | Ref. |
|---|---|---|---|---|---|---|---|---|
| Sprouty1 | X | (38) | ||||||
| Sprouty4 | X | X | (39) | |||||
| APC | X | (31, 32, 40) | ||||||
| CRBP1 | X | (32, 41) | ||||||
| CAV1 | X | (42, 43) | ||||||
| CCND2 | X | (32, 44, 45) | ||||||
| CD44 | X | (46-48) | ||||||
| TNFRSF10C | X | (49) | ||||||
| CDKN2A | X | X | (23, 32) | |||||
| DAPK | X | (50) | ||||||
| EDNRB | X | (51, 52) | ||||||
| ESR1 | X | (53-55) | ||||||
| ESR2 | X | (55-57) | ||||||
| FHIT | X | (58) | ||||||
| GSTPi | X | (40, 59, 60) | ||||||
| LPL | X | (61) | ||||||
| LAMA3 | X | (62) | ||||||
| LABM3 | X | (62) | ||||||
| MDR1 | (40) | |||||||
| PTGS1 | (63) | |||||||
| PTGS2 | (48, 53, 63) | |||||||
| RAR-β2 | (64) | |||||||
| RASSF1 | X | (65) | ||||||
| TIMP-3 | X | (12, 32) | ||||||
| TMS-1 | X | (41) | ||||||
| TGFBI | X | (66) | ||||||
| EphA7 | X | (67) | ||||||
| Sox7 | X | (68) | ||||||
| Smad4 | X | X | (69) | |||||
| HRK | X | (70) | ||||||
| GPX3 | X | (71) | ||||||
| TIMP-2 | X | (72) | ||||||
| LDHB | X | (73) | ||||||
| DLC-1 | X | (74) | ||||||
| PDLIM4 | X | (75) | ||||||
| CSR1 | X | X | (76) | |||||
| ALDH1a2 | X | (77) | ||||||
| DICE1 | X | (78) | ||||||
| TIG1 | X | (79, 80) |
HR=Hormone Receptor; CCC=Cell cycle control; RAD= Repair or avoidance of DNA damage; ST=Signal transduction; CBM=Cell adhesion and basement membrane; IR=Inflammation response; CTSG=candidate tumor suppressor Gene;
DNA METHYLATION ANALYSIS IN CLINICAL SAMPLES: CURRENTLY AVAILABLE METHODS
Differences in methylation patterns have also emerged as markers(81).. For example, differences between tumor and normal cells can be used to detect the presence of tumor cells in biopsy specimens or to identify tumor-derived DNA in blood samples (22, 23, 27-33, 53, 82-85). Differences in methylation patterns among tumors can be associated with patient outcome or other clinical responses, and can be used as markers to classify tumors. For these uses, methylation does not necessarily have to induce gene silencing, but simply be specific to tumor cells or have a pattern that is associated with clinically important information. Assays for methylation are appealing for translational research since they can utilize amplification techniques, such as methylation-specific polymerase chain reaction (PCR), and thereby utilize small amounts of samples. Due to its relative simplicity, safety, and sensitivity, methylation-specific PCR is the most commonly employed method for methylation analysis (86). Table 2 summarizes recent techniques that are evaluated in bodily fluids for prostate cancer detection. Table 3 showed recent findings of methylation of different gene or panel of genes as a prognostic marker. A brief outline of some the most promising techniques use for methylation analysis is described below:
Table 2.
List of DNA methylation assays tested in bodily fluids samples
| Method | Specimen | Gene/Gene Panel | Sensitivity | Specificity | Ref. |
|---|---|---|---|---|---|
| MSP | Urine | GSTP1 | 73 | 98 | (93) |
| MSP | Urine | GSTP1 | 79 | NR | (94) |
| QMSP | Urine | GSTP1,RASSF1a, RAR-β2, APC | 86 | 89 | (95) |
| QMSP | Urine | GSTP1 | 75 | 98 | (96) |
| QMSP | Urine | p16, ARF, MGMT, GSTP1 | 87 | 100 | (12) |
| Multiplexed QMSP |
Urine | GSTP1, RARB, APC | 55 | 80 | (97) |
| MSP | Serum | RASSF1, RARB2, GSTP1 | 28 | 100 | (98) |
| MSP | Serum | GSTP1 AR 14-3-3sigma |
32 40 87 |
100 73 45 |
(99) |
| QMSP | Serum |
MDR1 EDNRB RAR-β2 GTSP1 NEP RASSF1A |
83 50 39 28 17 17 |
100 100 100 100 100 100 |
(100) |
| MSRE QMSP |
Serum | GSTP1, TIG1, PTGS2, Reprimo | 42-47 | 92 | (101) |
MSP: Methylation specific PCR. It is gel-based, not highthroughput and subjective
QMSP: Quantitative-methylation specific PCR, automated, high-throughput and presumably more specific Mutiplexed PCR: High-throughput, cost-effective, can’t done more than 4 genes due to limitation in dye chemistry in real-time PCR. MSRE: methylation-sensitive restriction endonucleases; NR=not reported
Table 3.
Potentially useful methylation biomarkers for prognosis of prostate cancer
| Methods | Marker/Markers | Prognosis | Specimen | References |
|---|---|---|---|---|
| QMSP | PTGS2 | X | Primary tissue | (53) |
| MSP | GSTP1/APC GSTP1/PTGS2 GSTP1/APC/PTGS2 |
X | Primary tissue | (63) |
| MSP | CD44/ PTGS2 | X | Primary tissue | (48) |
| QMSP | APC | X | Primary tissue | (31) |
| Methylation Microarray and QMSP |
GPR7, ABHD9 and Chr3-EST |
X | Primary tissue | (102) |
| MethyLight | PTGS2, RAR-beta, and EDNRB |
X | Primary tissue | (103) |
| MSP | TMS1 | X | Primary tissue | (41) |
| QMSP | APC and cyclin D2 | X | Primary tissue | (45) |
| MSP | DRM, HPP1, RUNX3 |
X | Primary tissue | (104) |
| REQPCR | GTSP1 | X | serum | (105) |
| MSP | LAMA3 | X | Primary tissue | (62) |
| MSP | Cyclin D2 | X | Primary tissue | (44) |
| MSP | RASSF1A | X | Primary tissue | (65) |
Conventional Methylation-Specific PCR (MSP)
The conventional MSP assay uses two sets of primers specifically designed to amplify the methylated or unmethylated sequence, and the PCR products are run in a gel(86). Bisulfite-modified DNA are use as a template for PCR (86). The results of MSP at a particular DNA region are simply reported as methylated or unmethylated, not allowing quantitation or identification of partial methylation. The advantages of this method include a simple PCR based technique and can be perform any molecular biology laboratory, short time of analysis, the possibility of obtaining results from small amounts of DNA, specificity provided by primers and a significant sensitivity; methylation is detected even when only 0.1% of alleles are methylated. Additional drawbacks of this method, similarly to other PCR techniques, are connected with the possibility of contamination of the analyzed sample and obtaining false-positive results (86, 87). It is not automated and the interpretation of the data may be subjective.
MS-nested-PCR
It is a modified version of conventional MSP that involve a double-stage PCR (88). During the first step, the applied primers (primer with no CpG) recognized templates modified by sodium bisulfite, but they do not differentiate methylated sequences from unmethylated ones. Reaction products are diluted and amplified with two pairs of internal primers, one of them is specific for the methylated sequence and the other for the unmethylated one. The nested-PCR reaction increases the method’s sensitivity to 105, which made it possible to detect DNA methylation in saliva samples collected from patients with small cell lung cancer (89). Althoughnested-MSP are simple techniques that can easily be incorporated in most molecular biology laboratories, the ability to accurately determine the promoter methylation status of genes largely depends upon the careful design of MSP primers as well as other steps. The chances of contamination increase due to increasing number of PCR. This technique allow the analysis of individual (nested-MSP) or multiple (multiplex nested-MSP) promoters in samples with low quantity (e.g., macrodissected specimens) and/or quality (e.g., paraffin-embedded samples) (90, 91). This strategy is particularly attractive for assessing the methylation status of gene promoters in archival specimens for which clinical outcome (e.g., response to treatment, survival) is known (92) as well as for sensitive molecular detection (82). For prostate cancer these strategies can be useful for performing methylation profiling of large number of genes where only small of DNA are available like prostate intraepithelial neoplasia. This modified technique can also be use for the development of non-invasive assay (screening tool) using serum or urine DNA of high-risk man.
Quantitative analysis of methylated alleles (QAMA)
This technique is a newer version of Real-Time PCR, which applies TaqMan probes based on groove binder (MGB) technology. There are two types of probes: specific for methylated and unmethylated sequences, which is achieved by means of using different fluorescent dyes (VIC and FAM) (106, 107). The method is single stage, it ensures a high efficiency of methylation evaluation and the assessment of methylated and unmethylated sequences during one reaction, and it shortens significantly the time of analysis and diminishes the probability of contamination. Mutations and polymorphous sequences may hinder probes binding. The probes recognize only completely methylated or unmethylated sequences, which makes it impossible to analyze partially methylated areas (107).
Quantitative Methylation specific PCR (QMSP)
In this assay also bisulfite-modified DNA are use as a template for fluorescence-based real-time PCR, as previously described(12, 23, 25, 26) (108). QMSP is based on continuous monitoring of a progressive fluorogenic PCR by an optical system. Specific primers are designed to incorporate the CpG sites in order to detect the methylated form of the gene promoter, as described previously (86). QMSP is a variation which adds the use of a fluorescent probe across the CpG rich regions to add specificity (109). The probe is labeled with two fluorescent dyes: one serves as a reporter on the 5′-end (FAM), and its emission spectra is quenched by a second fluorescent dye on the 3′-end (TAMRA), when the two dyes are in close proximity. During the extension phase of the PCR, the 5′ to 3′ exonuclease activity of the Taq DNA polymerase cleaves the reporter from the probe, thus releasing it from the quencher, leading to fluorescence. If amplification occurs, this reaction results in an exponential increase in the fluorescent emission of the reporter dye and is monitored during the PCR process. β-actin (ACTB) is amplified in conjunction as a representative housekeeping gene to which the genes of interest are normalized, thus leading to a semi-quantitative result. This is important to ensure that there is a significant degree of methylation within sample, and not just trace amounts or contamination.
The Fluorogenic amplification reactions are carried out usually in triplicate in a volume of 20 μL that contained 3 μL of bisulfite-modified DNA; 600 nM concentrations of forward and reverse primers; 200 nM probe; 5 U of platinum Taq polymerase (Invitrogen); 200 μM concentrations each of dATP, dCTP, and dGTP; 200 μM dTTP; and 5.5 mM MgCl2. Fluorogenic probes were custom synthesized by PE Applied Biosystems. PCR primers were synthesized by Life Technologies (Gaithersburg, MD).Thermal cycling was initiated with a first denaturation step of 95° C for 10 min. The thermal profile for the PCR is 95°C for 15 s, 60° C(varies depending on gene and primer sequences) for 30 s and 72° C for 1 min. Data obtained following 50 cycles of amplification were analyzed. The later is just an example. Due to changing in technology and new enzyme and buffers, the PCR conditions are modified according to the investigator preference and the region of the genome to be amplified.
Amplification reactions were usually carried out in 384-well plates in a 7900 sequence detector (Perkin-Elmer Applied Biosystems) and are analyzed by a sequence detector system (SDS 2.2.1; Applied Biosystems). Each plate included patient DNA samples, positive (in vitro methylated leukocyte DNA) and negative (normal leukocyte DNA or DNA from a known unmethylated cell line) controls, and multiple water blanks. Leukocyte DNA from a healthy individual was methylated in vitro with excess SssI methyltransferase (New England Biolabs Inc., Beverly, MA) to generate completely methylated DNA. This enzyme adds methyl-groups to all CpG dinucleotides within the genomic DNA. Thus, in vitro methylated DNA is 100% positive for gene of interest promoter methylation after sodium-bisulfite treatment. Serial dilutions (90–0.009 ng) of this in vitro methylated DNA are used to construct a calibration curve for each plate. All samples should be within the assay’s range of sensitivity and reproducibility based on amplification of internal reference standard (threshold cycle [CT] value for ACTB of 40). The relative level of methylated DNA for each gene in each sample is determined as a ratio of methylation specific PCR-amplified gene to ACTB (reference gene) and then multiplied by 1000 for easier tabulation (average value of triplicates of gene of interest divided by the average value of triplicates of ACTB x 1000). The samples are categorized as unmethylated or methylated based on the sensitivity of the assay.
The QMSP assay provides several distinct advantages over conventional MSP: (a) omission of all of the post-amplification steps reduces the risk of contamination and increases the throughput of the system; (b) the assay is more stringent and more specific because in addition to the two PCR primers, the fluorescent-labeled hybridization probe has to anneal correctly between the two primers; (c) the assay is quantitative, automated, and readily adaptable to clinical setting and screening studies; and (d) the assay is amenable to multiplex amplification for the analysis of panels in clinical samples. At present, we can use four different dyes for the amplification of four distinct markers, but further developments in dye chemistry will improve the multi-marker diagnostic approach (presently, we can use four different dyes in Taqman technology) from nanogram quantities of low molecular weight DNA. These advances are unlikely to follow in conventional MSP.
Quantification of DNA methylation differences at specific sites using MS-SNuPE
Single-nucleotide primer extension is a well-established method which has been successfully used for the detection of gene mutations (110) and for the quantitation of allele specific expression (111-113). MSSNuPE relies on single nucleotide primer extension to assess DNA methylation at a specific cytosine (114). An initial round of PCR is carried out using bisulfite DNA-specific primers, followed by a second PCR step in which radio-labeled dCTP and dTTP and an internal primer which terminates precisely 5′ of the single nucleotide whose methylation status is to be determined are added. The radio-labeled products are then run on a 15% polyacrylamide gel under denaturing conditions and by visualized via exposure to an auto-radiographic film or a phosphor-image screen. The intensity of the observed bands can be then quantified to determine the proportion of C/T at the cytosine of interest. MS-SNuPE can be carried out in multiplex reactions, allowing for the quantification of more than a single CpG site per assay. MS-SNuPE is a viable alternative when sensitive quantitation of a single or few CpG sites is desired and small amounts of DNA are available.
Heavy-Methyl: PCR amplification of methylated DNA using methylation-specific oligonucleotide blockers
This is an innovative real-time variant of the MSPCR assay (86), which, because of its unique design, allows for the detection of methylated sequences at remarkably low concentration in a DNA mixture with high specificity (115). In this technique, the PCR priming is methylation specific, but the high specificity of the assay stems from the use of non-extendable oligo-nucleotide blockers. The blockers are designed to bind to the bisulfite-treated DNA template in a methylation-dependent manner and their binding sites are selected so as to overlap with the 3′ primer binding sites. Using primers specific for GSTP1, Heavy Methyl has been successfully used to detect 30 pg of in vitro methylated and bisulfite-treated DNA in a background of 50 ng unmethylated DNA (115). The high sensitivity of Heavy Methyl makes it suitable for clinical applications, such as the analysis of DNA methylation in serum, where the amount of non-cell bound free-floating DNA in healthy patients is estimated at 10–50 ng/ml (116, 117). An interesting feature of Heavy-Methyl is that it can be adapted for qualitative as well as quantitative analysis of DNA methylation. It is important to note that Heavy-Methyl requires more components and potentially more optimization than conventional MS-PCR, which has been used with high sensitivity and specificity for a large number of genes. Thus, Heavy-Methyl could provide an attractive technical alternative when convention MS-PCR is unsuitable for the goal of a given research endeavor.
Quantitative bisulfite sequencing using the pyrosequencing technology
Pyrosequencing is a sequence-by-synthesis approach that is based on the luminometric detection of pyrophosphate release following nucleotide incorporation (118, 119). Depending on the chemistry used, a three-to four-enzyme cascade converts the released pyrophosphate to ATP, which is immediately hydrolyzed to produce light. Since a single known nucleotide is added sequentially in each step, the obtained. One advantage of this method is that quantification can be achieved using SYBR Green I, which eliminates the need for fluorescently labeled probes, thus reducing the overall cost of the assay. Furthermore, given the widespread use of SYBR Green I in conventional real-time PCR assays, this technique could provide a suitable initial approach to DNA methylation analysis for researchers without prior experience in the DNA methylation field. However, as is the case with other PCR-based techniques described, careful primer design and optimization of the PCR reaction are critical to ensure the detection of the intended target sequence.
Prostate cancer-specific methylation markers in bodily fluids for early detection
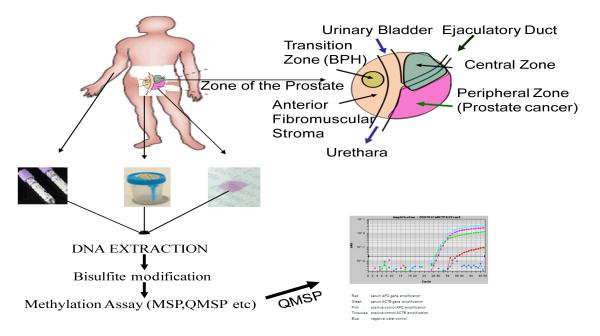
Cancer is a disease initiated and driven by the clonal evolution of cells transformed by genetic and epigenetic alterations, which can occur as either inherited (germline) mutations or acquired (somatic) mutations of key genes(120). Methylation, a epigenetic alterations (promoter methylation) can be used as targets for the detection of tumor cells in clinical specimens such as tissue biopsies or body fluids such as serum and plasma (Figure 1).
Figure 1. Methylation biomarkers for prostate cancer.
Tumor cells and free tumor DNA from dead cancer cells can access urine through secretion from the prostate into the urethra, and the proximity of urine to the transitional cell lining of the bladder and the renal system. Tumor cells that lack the capacity to metastasize and free tumor DNA can also access the circulatory system. Direct, but invasive, access by needle biopsy is performed for prostate cancer. DNA is isolated from the clinical specimen urine, blood or biopsy, and analysed for the presence of gene methylation byMSP or quantitative real-time methylation-specific PCR (qMSP).
SERUM
A potential cancer biomarker, cell-free circulating DNA, was already discovered in 1977 (121). Cancer patients have distinctly increased levels of cell-free DNA in comparison to healthy individuals and patients with non-malignant disorders (122). Although the assessment of cell-free DNA concentrations in serum/plasma is useful to identify patients with malignant disease, it was suggested that qualitative changes (i.e., presence of cancer-specific DNA alterations) provide the most accurate diagnostic information (123).
One of the earliest and most common somatic alterations in CaP is DNA CpG island (CGI) hypermethylation. For instance, CGI hypermethylation at the GSTP1 promoter was detected in approximately 80% of PCA tissues (124, 125). The use of two or more methylated genes even increased diagnostic accuracy (e.g. “GSTP1 and PTGS2”: sensitivity and specificity to 96–100%) (53, 63). Previous studies reported detection of hypermethylated DNA in serum and plasma of CaP patients: GSTP1 hypermethylation was detected in 12–30% of patients with metastatic disease (33, 105), and nearly in 100% of patients with metastatic PCA (83, 105). The presence of hypermethylated GSTP1 DNA fragments in serum was the strongest independent predictor of early PSA recurrence in patients undergoing radical prostatectomy for clinically localized CaP (105). Previous studies analyzing serum/plasma DNA focused on the detection of CGI hypermethylation at a single gene site in clinically localized CaP (33, 83, 105, 126). We are continuously discovering cancer-specific methylated gene by discovery approach (18). These genes are being tested for clinical utility such as diagnosis, prognosis etc. Specificity of methylation markers for diagnosis is depended on the compartment of the body. As for examples some methylation marker may be 100% specific for prostate cancer testing the serum. However, this marker may be terribly non-specific when urine is tested. Some methylation markers tested by our group and others have strong promise for developing screening tools using serum/plasma DNA.
URINE
As mentioned above, methylation of deoxycytidine residues within CpG islands in the upstream regulatory regions of a number of genes occurs in a very high percentage of prostate cancers and is not found to any significant extent in normal prostate tissues in most studies. Therefore, a number of groups (12, 33, 83, 84, 94, 127) have attempted to improve on the ability of serum PSA to predict a positive biopsy using methylation analysis of several cancer related genes in the urine and a number of these studies have been reviewed(128, 129). These studies have identified DNA methylation markers in the urine of men with prostate cancer as the basis for a confirmatorytest. Cairns et al.(94) reported that, when patients had methylation of GSTP1 detected in their tumor, 27% of cases were positive for DNA methylation in voided urine specimens. Jeronimo et al.(33) later showed sensitivity in urine of 23%. Gonzalgo et al.(84) demonstrated that GSTP1 methylation could be detected in the post-biopsy urine specimens of 39% of patients with prostate cancer. Rogers et al.(127) later demonstrated a high concordance between post-biopsy and post-DRE urinary samples for methylation of GSTP1, APC, and EDNRB. Goessl et al.(93) demonstrated a sensitivity of 73% at a specificity of 98% in urine sediments collected after prostate massage. Additional markers could potentially increase the sensitivity. For example, more recently, we found that a combination of CDKN2A (formerly p16), PSCD2 (formerly ARF), MGMT, and GSTP1 could theoretically enable the detection of 87% of prostate cancers at 100% specificity(12). In that study, GSTP1 alone demonstrated a sensitivity of 48% at a specificity of 100%. A study by Roupret et al.(95) examined GSTP1, RARB, APC, and RASSF1. The authors found a sensitivity of 86% at a specificity of 89%; however, they used a 1-min prostate massage and bladder catheterization, which would hinder widespread adoption.
One of the first studies using DNA-based tests was by Goessl et al(83)who used MSP to detect GSTP1 hypermethylation in bodily fluids. Although GSTP1 promoter hypermethylation was not detectable in prostate tissue and bodily fluids from patients with benign prostatic hyperplasia, these authors reported that methylation was detected in 94% of tumors (16 of 17), 72% of plasma or serum samples (23 of 32), 50% of ejaculate (4 of 8), and 36% of urine (4 of 11) from patients with prostate cancer. Additionally, MSP identified circulating tumor cells in 30% (10 of 33) of prostate cancer patients.
Goessl et al (93) also used MSP to detect GSTP1 hypermethylation in urine sediments from patients after prostate massage and found an overall sensitivity of 73% and a specificity of 98%, although some of these patients had advanced prostate cancer.
Roupre’t et al (95)recently used a 10 gene MSP approach in which urine samples were obtained from 95 consecutive radical prostatectomy patients and from 38 age-matched males (controls) with no history of genitourinary malignancy, negative prostate biopsies, and with or without benign prostatic hyperplasia. Radical prostatectomy patients underwent prostate massage and the first urine stream was then collected. The authors reported a sensitivity of 86% and a specificity of 89% for the 10 gene panel.
Results of another very recent study have been reported by Woodson et al(96) in which 100 men were referred for prostate needle biopsy due to increased PSA, abnormal digital rectal exam, or related symptoms. In this study, methylation of GSTP1 in post-massage urine had 75% sensitivity and 98% specificity for cancer. It is not clear why this latter study showed such high performance, but the results imply that perhaps the use of GSTP1 alone will be valuable as a molecular marker in prostate cancer in urine specimens.
Until now all of the studies that have detected methylation of genomic DNA in bodily fluids for the detection of prostate cancer have relied on some form of methylation-specific polymerase chain reaction (PCR). One major limitation of this approach is that the DNA must be first treated with sodium bisulfite, which is a very harsh treatment and results in damage to what are often already low quantities of DNA. Development of new kits for bisulfite treatment may overcome these issues. New technologies are also developing that can be use for detection of prostate cancer in bodily fluids DNA even without bisulfite treatment. One of such assay was recently developed by Yegnasubramanian et al(85). This assay, referred to as COMPARE-MS (combination methylated-DNA precipitation and methylation sensitive restriction enzymes) that does not rely on bisulfite treatment of DNA and this approach promises to increase the sensitivity of detecting CpG island hypermethylation. The approach, which results in very high sensitivity and specificity, features fragmenting genomic DNA with restriction enzymes including restriction enzymes that only cut when the target sequence is unmethylated, capture of methylated DNA using a purified recombinant methyl-binding domain polypeptide fragment from the human MBD2 protein, followed by PCR for the gene of interest. The assay was found to be highly sensitive and specific(85). It is anticipated that this type of approach may indeed improve upon existing approaches for both specific genes and for the ability to multiplex a number of genes.
Prostate cancer -specific methylation markers in primary tumors for the prediction of risk
During the past 15 years, widespread clinical use of the prostate-specific antigen (PSA) test has resulted in a significant increase in the proportion of patients presenting with early stages of disease. Currently, ~80% of newly diagnosed patients present with no clinical evidence of metastatic disease (130). Data from uncontrolled studies suggest that 30% to 50% of patients treated with local modalities will show evidence of biochemical (PSA) relapse within a 10-year follow-up period(5, 131-133). We evaluated the methylation status of the promoters of six genes, and their ability to add to known risk factors in predicting time to recurrence in prostate cancer patients following prostatectomy. These genes were chosen based on their ability to differentiate between benign hyperplasia of the prostate and prostate cancer (32). These genes were frequently found to be hypermethylated in prostate cancer and, in some studies, were related to certain clinicopathologic characteristics (58). Patient population was unique for two reasons. First, all patients had the same Gleason grade of 3 + 4 = 7. Second, none of the patients received any additional treatment after radical prostatectomy until metastatic disease was evident. This allowed us to evaluate the significance of hypermethylation in predicting aggressiveness of prostate cancer in a relatively homogeneous group of patients without interference in the natural biological history of the disease. A Gleason score of 7 was chosen because it represents the most heterogeneous group of patients in terms of outcome. Most patients with a Gleason score of ≤6 will be cured with radical prostatectomy only. In patients with a Gleason score of ≥8, the probability of recurrence is very high. In the multivariate analysis, including the most significant clinical and pathologic prognostic factors, hypermethylation of APC and a combined methylation profile of APC and cyclin D2 were significant predictors for TTP. The HR of combined APC and cyclin D2 hypermethylation was higher and more significant than the HR given by lymph node status or by the Kattan nomogram (134, 135). The latter combines the best-known clinical and pathologic risk factors for recurrence. These findings are especially significant because we evaluated a relatively homogenous group of patients with the same Gleason score.
In a recent study Cottrell et al.(102) discovered and validate 3 new DNA methylation markers for prostate cancer prognosis. They first used a genome-wide scan to identify new methylation targets and subsequently applied to a microarray of radical prostatectomy specimens to determine clinically relevant patterns of disease recurrence. Methylation levels of the 3 marker candidates GPR7, ABHD9 and an expressed sequence tag on chromosome 3 (Chr3-EST) were significantly increased in patients who did vs did not experience early PSA recurrence (Bonferroni correction p _0.05). Furthermore, these markers were also informative when the sample set was restricted to 68 mid range Gleason score (6 or 7) samples only. We developed real-time polymerase chain reaction assays for ABHD9 and Chr3-EST, and measured methylation in paraffin embedded, formalin fixed prostatectomy samples from an independent set of 223 patients. Methylation of the 2 markers was significantly higher in patients with early PSA recurrence compared to that in patients who did not experience PSA recurrence. They conclude that 3 novel markers GPR7, ABHD9 and Chr3-EST are significantly associated with prostate cancer prognosis. Incorporation of these methylation markers into clinical practice will result in more accurate prediction of which patients are likely to experience PSA recurrence. The next step ahead is to perform properly designed, prospective clinical trials to test new molecular markers if we hope to continue improving the outcomes of our patients. Only then the true usefulness of molecular markers for cancer diagnosis and prognosis are validated.
To understand the prognostic role of methylated genes in circulating cells DNA, Roupret et al.(136) tested ten gene promoter regions (GSTP1, RASSF1a, CDH1 (E-Cadherin), APC, DAPK, MGMT, p14, p16, RARb2 and TIMP3) in DNA extracted from whole blood for aberrant methylation by QMSP. They compared hypermethylation of circulating cell DNA from prostate cancer patients with (Group 1, n =20) and without (Group 2, n=22) disease progression and age-matched controls (benign prostatic hyperplasia, Group 3, n=22). They measured hypermethylation of 10 gene promoters in 2 sequential venous samples, obtained at diagnosis and during disease progression (median time, 15 months later). Matched time samples were obtained in the non-progressing patients. They found that more hypermethylation was detected in the diagnostic sample from the patients with cancer than in controls for GSTP1, RASSF1a, APC and RARβ2 (p < 0.0001). Patients undergoing disease progression had a significant increase in methylation levels of these 4 genes when compared to the other patients (p < 0.001). Patients at risk of disease progression have higher detectable concentrations of circulating cell hypermethylation, than those without progression. The extent of this hypermethylation increases during disease progression and they suggested that this approach can be used to identify the extent and duration of treatment response in prostate cancer. It is unclear why they targeted whole blood DNA for methylation study. Usually serum and plasma are the ideal source of DNA for methylation study for the detection and prognosis. Lymphocyte DNA in whole blood is not the source to test cancer related aberrant methylation. Tested DNA from all the three compartments (plasma, serum and lymphocyte) are needed to understand the specificity of their findings. If they found methylation in lymphocyte DNA, it can not be use as prognostic markers. However further study are essential to confirm their findings.
Expert Commentary
Methylation based markers have an enormous potential in (1) cancer risk assessment, (2) early detection, and (3) therapy monitoring. Numerous studies support the possible power of aberrant DNA-methylation for the development of screening tests to predict the individual risk for different types of cancer including prostate cancer. The identification of patients who are at high risk of cancer through screening could be very helpful for prophylactic treatment. The use of aberrant methylation as a cancer risk marker seems to be promising in recent years although multicenter blinded longitudinal studies are needed before using in a clinical setting. As a diagnostic marker, methylation marker has several advantages over protein, RNA or mutations markers. Firstly, the methylation signal can be amplified and therefore DNA methylation analysis by PCR is very sensitive although vigorous validation is needed to minimize false positive. Secondly, DNA is a very stable molecule in comparison to protein or RNA. In addition, DNA hypermethylation takes place in a defined area of the gene (usually in the “CpG island” of the promoter region), whereas mutations can take place in various regions of a gene that can be missed if total gene are not sequenced. For these reasons methylation analysis seems to be a promising tool in molecular diagnostics. In addition, paired normal is necessary for the exclusion of germline mutation while methylation markers if validated in age and gender matched normal, it is not necessary to test paired normal. For most types of cancer, early detection of the disease is associated with an improved clinical outcome. Because DNA methylation changes have been reported to occur early in carcinogenesis the identification of aberrant DNA methylation offers the exciting possibility of developing diagnostic tests. With a panel of prostate cancer specific methylated genes, urologists and pathologists have a tool of molecular diagnosis and a highly sensitive and specific non-invasive test not only for diagnosis but also for surveillance strategies. One advantage is the so-called multimarker strategy. Since one molecular marker fails to detect heterogeneous group of prostate cancers in a highly specific and sensitive manner, the methylation test (consist of panel of highly specific genes) examines panel of genes that includes all the important pathway genes that alter in prostate cancer. Quantitaive measurement of methylation at single base resolution will facilitate these endeavors. Technologies are now developing very rapidly for detecting methylation in a small amount of DNA. Multi-Plex PCR will allow including maximum number of genes in the diagnostic panel. This will also allow performing the test in a cost effective manner. The ongoing discussions about the usefulness of PSA as a marker for presence of prostate cancer can be overcome by addition of set of methylation markers in urine or serum. Presence of elevated PSA and promoter hypermethylation of certain genes in bodily fluids (urine, serum, prostate message fluids) may increase the specificity of the assay. As mentioned before, another advantage of DNA based screening test (like methylation test) is the stability of DNA, the bodily fluids can be collected by health workers and methylation markers can be tested in a centralize laboratory. So this screening strategy based on methylation markers may be useful for resource poor region. Analyzing methylation markers in serum can be use for monitoring of disease after complete prostatectomy.
Five-year view
For a biomarker to be clinically applicable, it must be specific, sensitive, and detectable in specimens obtained through minimally invasive procedures. Promising results have already been obtained since aberrantly methylated CpG islands have been detected in DNA samples derived from urine, serum, sputum, and stool of cancer patients. Of importance, it should be noted that changes in DNA methylation also occur in normal epithelia. Thus, extensive research is currently underway to identify tumor specific DNA methylation events that afford enough sensitivity and specificity to be utilized as biomarkers. Another major obstacle to overcome is the fact that tumor DNA is present only in minimal amounts in bodily fluids. Thus, exquisitely sensitive techniques need to be utilized to detect and analyze tumor-derived DNA.
There are currently many different approaches to generate DNA methylation data. A large number of these are well-established and have been important tools for epigenetic analysis for many years. However, no single technique provides an unambiguous approach to DNA methylation data harvesting. Thus, I have tried to provide a description of the advantages and disadvantages of various techniques, in an attempt to provide a framework useful when deciding which method to use to generate the most meaningful data.
I would like to emphasize the critical role of DNA methylation assays as tools for the assessment of the effectiveness and safety of DNA demethylating agents, as they potentially develop into standard regiments for cancer therapy. Drugs such as decitabine have shown promising results in clinical trials focused on the treatment of solid and liquid tumors. However, due to the nonspecific nature of nucleotide analogs, it is critical to monitor their effect not only on neoplastic cells but also on normal tissues to ensure that no long-term damage is inflicted to unaffected targets.A large body of evidence now exists indicating that not all possible DNA methylation targets in the human genome are affected equally in the disease state. The biological mechanism behind these observations is currently not fully understood, but could involve selection pressure or an intrinsic difference in sequence susceptibility to aberrant epigenetic changes. Thus, the use of sensitive assays to monitor DNA methylation changes will play a key role in the development and implementation of new therapies aimed at modulating the epigenome.
Key issues.
Even with prostate-specific antigen (PSA) testing, there is significant benefit to be gained from new methods for the early detection of prostate cancer.
Aberrant hypermethylation of the promoter region of genes is a frequent and early event in cancer cells. The hypermethylation is associated with loss of function of the gene.
Epigenetic silencing can be reversed using small molecule inhibitors of methylation and deacetylation, some of which are either US FDA approved or currently in clinical trials.
Highly sensitive and specific DNA based assay like DNA-methylation-specific PCR technology allow the detection of gene methylation from rare tumor cells in tissue biopsies and bodily fluids (eg, urine, serum etc). Conceptually, the methylation of a set of tumor-suppressor genes is highly specific for neoplastic cells.
Several studies have demonstrated sensitive and specific detection of gene methylation in urine and serum from patients with early stage prostate.
Clinical materials suitable for DNA methylation assays is readily accessible from patients of prostate cancer and may prove useful for screening, diagnosis, prognostication.
One of the main challenges to the clinical implementation of gene methylation-based detection include the need for validation in larger, well-defined populations with optimized and standardized methodology in multi-centered setting. Further insight into the timing of gene methylation during the earliest stages of neoplastic development will be important.
Genome-wide profiling efforts may facilitate for identification of more and better epigenetic markers useful for prostate cancer management with improved sensitivity and specificity.
Age-dependent changes in methylation have been found in several genes relevant to cancer and suggest one explanation for the rising prevalence of cancer in the elderly.
Future directions will probably involve screening with gene methylation for the simultaneous detection, differential diagnosis and prediction of future behaviour of prostate cancer in a single non-invasive body fluid specimen. Surveillance for the early detection of recurrent tumor may also be perform by methylation assay.
Acknowledgments
Grant support: Flight Attendant Medical Research Institute Young Clinical Scientist Award, International Association for the Study of Lung and Career development award from SPORE in Cervical Cancer Grants P50 CA098252 (M.O. Hoque); Dr. Hoque is a paid consultant to Oncomethylome Sciences, SA.
REFERENCES
Papers of special note have been highlighted as: ° of interest °° of considerable interest
- 1.Quinn DI, Henshall SM, Sutherland RL. Molecular markers of prostate cancer outcome. Eur J Cancer. 2005;41:858–887. doi: 10.1016/j.ejca.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66. doi: 10.3322/canjclin.57.1.43. [DOI] [PubMed] [Google Scholar]
- 3.Yao SL, Lu-Yao G. Understanding and appreciating overdiagnosis in the PSA era. J Natl Cancer Inst. 2002;94:958–960. doi: 10.1093/jnci/94.13.958. [DOI] [PubMed] [Google Scholar]
- 4.Alers JC, Krijtenburg PJ, Vis AN, Hoedemaeker RF, Wildhagen MF, Hop WC, van Der Kwast TT, Schroder FH, Tanke HJ, van Dekken H. Molecular cytogenetic analysis of prostatic adenocarcinomas from screening studies : early cancers may contain aggressive genetic features. Am J Pathol. 2001;158:399–406. doi: 10.1016/s0002-9440(10)63983-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Han M, Partin AW, Zahurak M, Piantadosi S, Epstein JI, Walsh PC. Biochemical (prostate specific antigen) recurrence probability following radical prostatectomy for clinically localized prostate cancer. J Urol. 2003;169:517–523. doi: 10.1097/01.ju.0000045749.90353.c7. [DOI] [PubMed] [Google Scholar]
- 6.Barratt AL, Coates AS. Screening decreases prostate cancer death: first analysis of the 1988 Quebec Prospective Randomized Controlled Trial. Med J Aust. 2004;181:213–214. doi: 10.5694/j.1326-5377.2004.tb06240.x. [DOI] [PubMed] [Google Scholar]
- 7.Labrie F, Candas B, Dupont A, Cusan L, Gomez JL, Suburu RE, Diamond P, Levesque J, Belanger A. Screening decreases prostate cancer death: first analysis of the 1988 Quebec prospective randomized controlled trial. Prostate. 1999;38:83–91. doi: 10.1002/(sici)1097-0045(19990201)38:2<83::aid-pros1>3.0.co;2-b. [DOI] [PubMed] [Google Scholar]
- 8.Baylin SB, Herman JG, Graff JR, Vertino PM, Issa JP. Alterations in DNA methylation: a fundamental aspect of neoplasia. Adv Cancer Res. 1998;72:141–196. [PubMed] [Google Scholar]
- 9.Bird A. The essentials of DNA methylation. Cell. 1992;70:5–8. doi: 10.1016/0092-8674(92)90526-i. [DOI] [PubMed] [Google Scholar]
- 10.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 11.Herman JG, Umar A, Polyak K, Graff JR, Ahuja N, Issa JP, Markowitz S, Willson JK, Hamilton SR, Kinzler KW, Kane MF, Kolodner RD, Vogelstein B, Kunkel TA, Baylin SB. Incidence and functional consequences of hMLH1 promoter hypermethylation in colorectal carcinoma. Proc Natl Acad Sci U S A. 1998;95:6870–6875. doi: 10.1073/pnas.95.12.6870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoque MO, Topaloglu O, Begum S, Henrique R, Rosenbaum E, Van Criekinge W, Westra WH, Sidransky D. Quantitative methylation-specific polymerase chain reaction gene patterns in urine sediment distinguish prostate cancer patients from control subjects. J Clin Oncol. 2005;23:6569–6575. doi: 10.1200/JCO.2005.07.009. °°This study demonstrated the sensitivity and specificity of panel of methylated gene by quantitative-methylation specific PCR for the diagnosis of prostate cancer
- 13.Cooper CS, Foster CS. Concepts of epigenetics in prostate cancer development. Br J Cancer. 2009;100:240–245. doi: 10.1038/sj.bjc.6604771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nelson WG, Yegnasubramanian S, Agoston AT, Bastian PJ, Lee BH, Nakayama M, De Marzo AM. Abnormal DNA methylation, epigenetics, and prostate cancer. Front Biosci. 2007;12:4254–4266. doi: 10.2741/2385. [DOI] [PubMed] [Google Scholar]
- 15.Dobosy JR, Roberts JL, Fu VX, Jarrard DF. The expanding role of epigenetics in the development, diagnosis and treatment of prostate cancer and benign prostatic hyperplasia. J Urol. 2007;177:822–831. doi: 10.1016/j.juro.2006.10.063. [DOI] [PubMed] [Google Scholar]
- 16.Li LC. Epigenetics of prostate cancer. Front Biosci. 2007;12:3377–3397. doi: 10.2741/2320. [DOI] [PubMed] [Google Scholar]
- 17.Esteller M. Cancer epigenomics: DNA methylomes and histone-modification maps. Nat Rev Genet. 2007;8:286–298. doi: 10.1038/nrg2005. [DOI] [PubMed] [Google Scholar]
- 18.Hoque MO, Kim MS, Ostrow KL, Liu J, Wisman GB, Park HL, Poeta ML, Jeronimo C, Henrique R, Lendvai A, Schuuring E, Begum S, Rosenbaum E, Ongenaert M, Yamashita K, Califano J, Westra W, van der Zee AG, Van Criekinge W, Sidransky D. Genome-wide promoter analysis uncovers portions of the cancer methylome. Cancer Res. 2008;68:2661–2670. doi: 10.1158/0008-5472.CAN-07-5913. °°This is a modified and greatly improved approach for the selection of candidate methylated genes based on new promoter structure algorithm and microarray data generated from 20 cancer cell lines of 5 major cancer types including prostate cancer.
- 19.Ngan RK, Lau WH, Yip TT, Cho WC, Cheng WW, Lim CK, Wan KK, Chu E, Joab I, Grunewald V, Poon YF, Ho JH. Remarkable application of serum EBV EBER-1 in monitoring response of nasopharyngeal cancer patients to salvage chemotherapy. Ann N Y Acad Sci. 2001;945:73–79. doi: 10.1111/j.1749-6632.2001.tb03866.x. [DOI] [PubMed] [Google Scholar]
- 20.Lo YM. Prognostic implication of pretreatment plasma/serum concentration of Epstein-Barr virus DNA in nasopharyngeal carcinoma. Biomed Pharmacother. 2001;55:362–365. doi: 10.1016/s0753-3322(01)00083-x. [DOI] [PubMed] [Google Scholar]
- 21.Sidransky D, Von Eschenbach A, Tsai YC, Jones P, Summerhayes I, Marshall F, Paul M, Green P, Hamilton SR, Frost P, et al. Identification of p53 gene mutations in bladder cancers and urine samples. Science. 1991;252:706–709. doi: 10.1126/science.2024123. [DOI] [PubMed] [Google Scholar]
- 22.Cairns P. Gene methylation and early detection of genitourinary cancer: the road ahead. Nat Rev Cancer. 2007;7:531–543. doi: 10.1038/nrc2170. °°Recent systemic review on methylation as a biomarker for genetourinary cancer
- 23.Hoque MO, Begum S, Topaloglu O, Jeronimo C, Mambo E, Westra WH, Califano JA, Sidransky D. Quantitative detection of promoter hypermethylation of multiple genes in the tumor, urine, and serum DNA of patients with renal cancer. Cancer Res. 2004;64:5511–5517. doi: 10.1158/0008-5472.CAN-04-0799. [DOI] [PubMed] [Google Scholar]
- 24.Topaloglu O, Hoque MO, Tokumaru Y, Lee J, Ratovitski E, Sidransky D, Moon CS. Detection of promoter hypermethylation of multiple genes in the tumor and bronchoalveolar lavage of patients with lung cancer. Clin Cancer Res. 2004;10:2284–2288. doi: 10.1158/1078-0432.ccr-1111-3. [DOI] [PubMed] [Google Scholar]
- 25.Hoque MO, Begum S, Topaloglu O, Chatterjee A, Rosenbaum E, Van Criekinge W, Westra WH, Schoenberg M, Zahurak M, Goodman SN, Sidransky D. Quantitation of promoter methylation of multiple genes in urine DNA and bladder cancer detection. J Natl Cancer Inst. 2006;98:996–1004. doi: 10.1093/jnci/djj265. [DOI] [PubMed] [Google Scholar]
- 26.Hoque MO, Feng Q, Toure P, Dem A, Critchlow CW, Hawes SE, Wood T, Jeronimo C, Rosenbaum E, Stern J, Yu M, Trink B, Kiviat NB, Sidransky D. Detection of aberrant methylation of four genes in plasma DNA for the detection of breast cancer. J Clin Oncol. 2006;24:4262–4269. doi: 10.1200/JCO.2005.01.3516. [DOI] [PubMed] [Google Scholar]
- 27.Henrique R, Costa VL, Cerveira N, Carvalho AL, Hoque MO, Ribeiro FR, Oliveira J, Teixeira MR, Sidransky D, Jeronimo C. Hypermethylation of Cyclin D2 is associated with loss of mRNA expression and tumor development in prostate cancer. J Mol Med. 2006;84:911–918. doi: 10.1007/s00109-006-0099-4. [DOI] [PubMed] [Google Scholar]
- 28.Henrique R, Jeronimo C, Hoque MO, Carvalho AL, Oliveira J, Teixeira MR, Lopes C, Sidransky D. Frequent 14-3-3 sigma promoter methylation in benign and malignant prostate lesions. DNA Cell Biol. 2005;24:264–269. doi: 10.1089/dna.2005.24.264. [DOI] [PubMed] [Google Scholar]
- 29.Henrique R, Jeronimo C, Hoque MO, Nomoto S, Carvalho AL, Costa VL, Oliveira J, Teixeira MR, Lopes C, Sidransky D. MT1G hypermethylation is associated with higher tumor stage in prostate cancer. Cancer Epidemiol Biomarkers Prev. 2005;14:1274–1278. doi: 10.1158/1055-9965.EPI-04-0659. [DOI] [PubMed] [Google Scholar]
- 30.Henrique R, Jeronimo C, Teixeira MR, Hoque MO, Carvalho AL, Pais I, Ribeiro FR, Oliveira J, Lopes C, Sidransky D. Epigenetic heterogeneity of high-grade prostatic intraepithelial neoplasia: clues for clonal progression in prostate carcinogenesis. Mol Cancer Res. 2006;4:1–8. doi: 10.1158/1541-7786.MCR-05-0113. [DOI] [PubMed] [Google Scholar]
- 31.Henrique R, Ribeiro FR, Fonseca D, Hoque MO, Carvalho AL, Costa VL, Pinto M, Oliveira J, Teixeira MR, Sidransky D, Jeronimo C. High promoter methylation levels of APC predict poor prognosis in sextant biopsies from prostate cancer patients. Clin Cancer Res. 2007;13:6122–6129. doi: 10.1158/1078-0432.CCR-07-1042. °A key report of APC gene methylation and poor prognosis in prostate cancer
- 32.Jeronimo C, Henrique R, Hoque MO, Mambo E, Ribeiro FR, Varzim G, Oliveira J, Teixeira MR, Lopes C, Sidransky D. A quantitative promoter methylation profile of prostate cancer. Clin Cancer Res. 2004;10:8472–8478. doi: 10.1158/1078-0432.CCR-04-0894. ° A comprehensive methylation profiling of prostate cancer by candidate gene approach
- 33.Jeronimo C, Usadel H, Henrique R, Silva C, Oliveira J, Lopes C, Sidransky D. Quantitative GSTP1 hypermethylation in bodily fluids of patients with prostate cancer. Urology. 2002;60:1131–1135. doi: 10.1016/s0090-4295(02)01949-0. [DOI] [PubMed] [Google Scholar]
- 34.Nakayama M, Bennett CJ, Hicks JL, Epstein JI, Platz EA, Nelson WG, De Marzo AM. Hypermethylation of the human glutathione S-transferase-pi gene (GSTP1) CpG island is present in a subset of proliferative inflammatory atrophy lesions but not in normal or hyperplastic epithelium of the prostate: a detailed study using laser-capture microdissection. Am J Pathol. 2003;163:923–933. doi: 10.1016/s0002-9440(10)63452-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lombaerts M, van Wezel T, Philippo K, Dierssen JW, Zimmerman RM, Oosting J, van Eijk R, Eilers PH, van de Water B, Cornelisse CJ, Cleton-Jansen AM. E-cadherin transcriptional downregulation by promoter methylation but not mutation is related to epithelial-to-mesenchymal transition in breast cancer cell lines. Br J Cancer. 2006;94:661–671. doi: 10.1038/sj.bjc.6602996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li LC, Shiina H, Deguchi M, Zhao H, Okino ST, Kane CJ, Carroll PR, Igawa M, Dahiya R. Age-dependent methylation of ESR1 gene in prostate cancer. Biochem Biophys Res Commun. 2004;321:455–461. doi: 10.1016/j.bbrc.2004.06.164. [DOI] [PubMed] [Google Scholar]
- 37.Wang Q, Williamson M, Bott S, Brookman-Amissah N, Freeman A, Nariculam J, Hubank MJ, Ahmed A, Masters JR. Hypomethylation of WNT5A, CRIP1 and S100P in prostate cancer. Oncogene. 2007;26:6560–6565. doi: 10.1038/sj.onc.1210472. [DOI] [PubMed] [Google Scholar]
- 38.Kwabi-Addo B, Ren C, Ittmann M. DNA methylation and aberrant expression of sprouty1 in human prostate cancer. Epigenetics. 2009;4 doi: 10.4161/epi.4.1.7400. [DOI] [PubMed] [Google Scholar]
- 39.Wang J, Thompson B, Ren C, Ittmann M, Kwabi-Addo B. Sprouty4, a suppressor of tumor cell motility, is down regulated by DNA methylation in human prostate cancer. Prostate. 2006;66:613–624. doi: 10.1002/pros.20353. [DOI] [PubMed] [Google Scholar]
- 40.Enokida H, Shiina H, Urakami S, Igawa M, Ogishima T, Li LC, Kawahara M, Nakagawa M, Kane CJ, Carroll PR, Dahiya R. Multigene methylation analysis for detection and staging of prostate cancer. Clin Cancer Res. 2005;11:6582–6588. doi: 10.1158/1078-0432.CCR-05-0658. °This is the first report demonstrating that M score is a new method for multigene methylation analysis that can serve as a good diagnostic and staging biomarker for prostate cancer.
- 41.Suzuki M, Shigematsu H, Shivapurkar N, Reddy J, Miyajima K, Takahashi T, Gazdar AF, Frenkel EP. Methylation of apoptosis related genes in the pathogenesis and prognosis of prostate cancer. Cancer Lett. 2006;242:222–230. doi: 10.1016/j.canlet.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 42.Bachmann N, Haeusler J, Luedeke M, Kuefer R, Perner S, Assum G, Paiss T, Hoegel J, Vogel W, Maier C. Expression changes of CAV1 and EZH2, located on 7q31 approximately q36, are rarely related to genomic alterations in primary prostate carcinoma. Cancer Genet Cytogenet. 2008;182:103–110. doi: 10.1016/j.cancergencyto.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 43.Cui J, Rohr LR, Swanson G, Speights VO, Maxwell T, Brothman AR. Hypermethylation of the caveolin-1 gene promoter in prostate cancer. Prostate. 2001;46:249–256. doi: 10.1002/1097-0045(20010215)46:3<249::aid-pros1030>3.0.co;2-#. [DOI] [PubMed] [Google Scholar]
- 44.Padar A, Sathyanarayana UG, Suzuki M, Maruyama R, Hsieh JT, Frenkel EP, Minna JD, Gazdar AF. Inactivation of cyclin D2 gene in prostate cancers by aberrant promoter methylation. Clin Cancer Res. 2003;9:4730–4734. [PubMed] [Google Scholar]
- 45.Rosenbaum E, Hoque MO, Cohen Y, Zahurak M, Eisenberger MA, Epstein JI, Partin AW, Sidransky D. Promoter hypermethylation as an independent prognostic factor for relapse in patients with prostate cancer following radical prostatectomy. Clin Cancer Res. 2005;11:8321–8325. doi: 10.1158/1078-0432.CCR-05-1183. °°This is the first study demonstrating that methylation status of selected genes in the prostate cancer specimen may predict for time to recurrence in Gleason 3 + 4 = 7 patients undergoing prostatectomy. These results should be validated in a larger and unselected cohort.
- 46.Hanson JA, Gillespie JW, Grover A, Tangrea MA, Chuaqui RF, Emmert-Buck MR, Tangrea JA, Libutti SK, Linehan WM, Woodson KG. Gene promoter methylation in prostate tumor-associated stromal cells. J Natl Cancer Inst. 2006;98:255–261. doi: 10.1093/jnci/djj051. [DOI] [PubMed] [Google Scholar]
- 47.Woodson K, Hayes R, Wideroff L, Villaruz L, Tangrea J. Hypermethylation of GSTP1, CD44, and E-cadherin genes in prostate cancer among US Blacks and Whites. Prostate. 2003;55:199–205. doi: 10.1002/pros.10236. [DOI] [PubMed] [Google Scholar]
- 48.Woodson K, O’Reilly KJ, Ward DE, Walter J, Hanson J, Walk EL, Tangrea JA. CD44 and PTGS2 methylation are independent prognostic markers for biochemical recurrence among prostate cancer patients with clinically localized disease. Epigenetics. 2006;1:183–186. doi: 10.4161/epi.1.4.3530. [DOI] [PubMed] [Google Scholar]
- 49.Cheng Y, Kim JW, Liu W, Dunn TA, Luo J, Loza MJ, Kim ST, Zheng SL, Xu J, Isaacs WB, Chang BL. Genetic and epigenetic inactivation of TNFRSF10C in human prostate cancer. Prostate. 2009;69:327–335. doi: 10.1002/pros.20882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yamanaka M, Watanabe M, Yamada Y, Takagi A, Murata T, Takahashi H, Suzuki H, Ito H, Tsukino H, Katoh T, Sugimura Y, Shiraishi T. Altered methylation of multiple genes in carcinogenesis of the prostate. Int J Cancer. 2003;106:382–387. doi: 10.1002/ijc.11227. [DOI] [PubMed] [Google Scholar]
- 51.Nelson JB, Lee WH, Nguyen SH, Jarrard DF, Brooks JD, Magnuson SR, Opgenorth TJ, Nelson WG, Bova GS. Methylation of the 5′ CpG island of the endothelin B receptor gene is common in human prostate cancer. Cancer Res. 1997;57:35–37. [PubMed] [Google Scholar]
- 52.Jeronimo C, Henrique R, Campos PF, Oliveira J, Caballero OL, Lopes C, Sidransky D. Endothelin B receptor gene hypermethylation in prostate adenocarcinoma. J Clin Pathol. 2003;56:52–55. doi: 10.1136/jcp.56.1.52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yegnasubramanian S, Kowalski J, Gonzalgo ML, Zahurak M, Piantadosi S, Walsh PC, Bova GS, De Marzo AM, Isaacs WB, Nelson WG. Hypermethylation of CpG islands in primary and metastatic human prostate cancer. Cancer Res. 2004;64:1975–1986. doi: 10.1158/0008-5472.can-03-3972. [DOI] [PubMed] [Google Scholar]
- 54.Moriyama-Gonda N, Shiina H, Terashima M, Satoh K, Igawa M. Rationale and clinical implication of combined chemotherapy with cisplatin and oestrogen in prostate cancer: primary evidence based on methylation analysis of oestrogen receptor-alpha. BJU Int. 2008;101:485–491. doi: 10.1111/j.1464-410X.2007.07256.x. [DOI] [PubMed] [Google Scholar]
- 55.Sasaki M, Tanaka Y, Perinchery G, Dharia A, Kotcherguina I, Fujimoto S, Dahiya R. Methylation and inactivation of estrogen, progesterone, and androgen receptors in prostate cancer. J Natl Cancer Inst. 2002;94:384–390. doi: 10.1093/jnci/94.5.384. [DOI] [PubMed] [Google Scholar]
- 56.Lau KM, LaSpina M, Long J, Ho SM. Expression of estrogen receptor (ER)-alpha and ER-beta in normal and malignant prostatic epithelial cells: regulation by methylation and involvement in growth regulation. Cancer Res. 2000;60:3175–3182. [PubMed] [Google Scholar]
- 57.Zhu X, Leav I, Leung YK, Wu M, Liu Q, Gao Y, McNeal JE, Ho SM. Dynamic regulation of estrogen receptor-beta expression by DNA methylation during prostate cancer development and metastasis. Am J Pathol. 2004;164:2003–2012. doi: 10.1016/s0002-9440(10)63760-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Maruyama R, Toyooka S, Toyooka KO, Virmani AK, Zochbauer-Muller S, Farinas AJ, Minna JD, McConnell J, Frenkel EP, Gazdar AF. Aberrant promoter methylation profile of prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2002;8:514–519. [PubMed] [Google Scholar]
- 59.Lee WH, Morton RA, Epstein JI, Brooks JD, Campbell PA, Bova GS, Hsieh WS, Isaacs WB, Nelson WG. Cytidine methylation of regulatory sequences near the pi-class glutathione S-transferase gene accompanies human prostatic carcinogenesis. Proc Natl Acad Sci U S A. 1994;91:11733–11737. doi: 10.1073/pnas.91.24.11733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jeronimo C, Usadel H, Henrique R, Oliveira J, Lopes C, Nelson WG, Sidransky D. Quantitation of GSTP1 methylation in non-neoplastic prostatic tissue and organ-confined prostate adenocarcinoma. J Natl Cancer Inst. 2001;93:1747–1752. doi: 10.1093/jnci/93.22.1747. [DOI] [PubMed] [Google Scholar]
- 61.Kim JW, Cheng Y, Liu W, Li T, Yegnasubramanian S, Zheng SL, Xu J, Isaacs WB, Chang BL. Genetic and epigenetic inactivation of LPL gene in human prostate cancer. Int J Cancer. 2009;124:734–738. doi: 10.1002/ijc.23972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sathyanarayana UG, Padar A, Suzuki M, Maruyama R, Shigematsu H, Hsieh JT, Frenkel EP, Gazdar AF. Aberrant promoter methylation of laminin-5-encoding genes in prostate cancers and its relationship to clinicopathological features. Clin Cancer Res. 2003;9:6395–6400. [PubMed] [Google Scholar]
- 63.Bastian PJ, Ellinger J, Wellmann A, Wernert N, Heukamp LC, Muller SC, von Ruecker A. Diagnostic and prognostic information in prostate cancer with the help of a small set of hypermethylated gene loci. Clin Cancer Res. 2005;11:4097–4106. doi: 10.1158/1078-0432.CCR-04-1832. [DOI] [PubMed] [Google Scholar]
- 64.Jeronimo C, Henrique R, Hoque MO, Ribeiro FR, Oliveira J, Fonseca D, Teixeira MR, Lopes C, Sidransky D. Quantitative RARbeta2 hypermethylation: a promising prostate cancer marker. Clin Cancer Res. 2004;10:4010–4014. doi: 10.1158/1078-0432.CCR-03-0643. [DOI] [PubMed] [Google Scholar]
- 65.Liu L, Yoon JH, Dammann R, Pfeifer GP. Frequent hypermethylation of the RASSF1A gene in prostate cancer. Oncogene. 2002;21:6835–6840. doi: 10.1038/sj.onc.1205814. [DOI] [PubMed] [Google Scholar]
- 66.Shah JN, Shao G, Hei TK, Zhao Y. Methylation screening of the TGFBI promoter in human lung and prostate cancer by methylation-specific PCR. BMC Cancer. 2008;8:284. doi: 10.1186/1471-2407-8-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Guan M, Xu C, Zhang F, Ye C. Aberrant methylation of EphA7 in human prostate cancer and its relation to clinicopathologic features. Int J Cancer. 2009;124:88–94. doi: 10.1002/ijc.23890. [DOI] [PubMed] [Google Scholar]
- 68.Guo L, Zhong D, Lau S, Liu X, Dong XY, Sun X, Yang VW, Vertino PM, Moreno CS, Varma V, Dong JT, Zhou W. Sox7 Is an independent checkpoint for beta-catenin function in prostate and colon epithelial cells. Mol Cancer Res. 2008;6:1421–1430. doi: 10.1158/1541-7786.MCR-07-2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Aitchison AA, Veerakumarasivam A, Vias M, Kumar R, Hamdy FC, Neal DE, Mills IG. Promoter methylation correlates with reduced Smad4 expression in advanced prostate cancer. Prostate. 2008;68:661–674. doi: 10.1002/pros.20730. [DOI] [PubMed] [Google Scholar]
- 70.Higuchi T, Nakamura M, Shimada K, Ishida E, Hirao K, Konishi N. HRK inactivation associated with promoter methylation and LOH in prostate cancer. Prostate. 2008;68:105–113. doi: 10.1002/pros.20600. [DOI] [PubMed] [Google Scholar]
- 71.Yu YP, Yu G, Tseng G, Cieply K, Nelson J, Defrances M, Zarnegar R, Michalopoulos G, Luo JH. Glutathione peroxidase 3, deleted or methylated in prostate cancer, suppresses prostate cancer growth and metastasis. Cancer Res. 2007;67:8043–8050. doi: 10.1158/0008-5472.CAN-07-0648. [DOI] [PubMed] [Google Scholar]
- 72.Pulukuri SM, Patibandla S, Patel J, Estes N, Rao JS. Epigenetic inactivation of the tissue inhibitor of metalloproteinase-2 (TIMP-2) gene in human prostate tumors. Oncogene. 2007;26:5229–5237. doi: 10.1038/sj.onc.1210329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Leiblich A, Cross SS, Catto JW, Phillips JT, Leung HY, Hamdy FC, Rehman I. Lactate dehydrogenase-B is silenced by promoter hypermethylation in human prostate cancer. Oncogene. 2006;25:2953–2960. doi: 10.1038/sj.onc.1209262. [DOI] [PubMed] [Google Scholar]
- 74.Guan M, Zhou X, Soulitzis N, Spandidos DA, Popescu NC. Aberrant methylation and deacetylation of deleted in liver cancer-1 gene in prostate cancer: potential clinical applications. Clin Cancer Res. 2006;12:1412–1419. doi: 10.1158/1078-0432.CCR-05-1906. [DOI] [PubMed] [Google Scholar]
- 75.Vanaja DK, Ballman KV, Morlan BW, Cheville JC, Neumann RM, Lieber MM, Tindall DJ, Young CY. PDLIM4 repression by hypermethylation as a potential biomarker for prostate cancer. Clin Cancer Res. 2006;12:1128–1136. doi: 10.1158/1078-0432.CCR-05-2072. [DOI] [PubMed] [Google Scholar]
- 76.Yu G, Tseng GC, Yu YP, Gavel T, Nelson J, Wells A, Michalopoulos G, Kokkinakis D, Luo JH. CSR1 suppresses tumor growth and metastasis of prostate cancer. Am J Pathol. 2006;168:597–607. doi: 10.2353/ajpath.2006.050620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kim H, Lapointe J, Kaygusuz G, Ong DE, Li C, van de Rijn M, Brooks JD, Pollack JR. The retinoic acid synthesis gene ALDH1a2 is a candidate tumor suppressor in prostate cancer. Cancer Res. 2005;65:8118–8124. doi: 10.1158/0008-5472.CAN-04-4562. [DOI] [PubMed] [Google Scholar]
- 78.Ropke A, Buhtz P, Bohm M, Seger J, Wieland I, Allhoff EP, Wieacker PF. Promoter CpG hypermethylation and downregulation of DICE1 expression in prostate cancer. Oncogene. 2005;24:6667–6675. doi: 10.1038/sj.onc.1208824. [DOI] [PubMed] [Google Scholar]
- 79.Zhang J, Liu L, Pfeifer GP. Methylation of the retinoid response gene TIG1 in prostate cancer correlates with methylation of the retinoic acid receptor beta gene. Oncogene. 2004;23:2241–2249. doi: 10.1038/sj.onc.1207328. [DOI] [PubMed] [Google Scholar]
- 80.Tokumaru Y, Sun DI, Nomoto S, Yamashita K, Sidransky D. Re: Is TIG1 a new tumor suppressor in prostate cancer? J Natl Cancer Inst. 2003;95:919–920. doi: 10.1093/jnci/95.12.919. [DOI] [PubMed] [Google Scholar]
- 81.Verma M, Srivastava S. Epigenetics in cancer: implications for early detection and prevention. Lancet Oncol. 2002;3:755–763. doi: 10.1016/s1470-2045(02)00932-4. [DOI] [PubMed] [Google Scholar]
- 82.Belinsky SA, Liechty KC, Gentry FD, Wolf HJ, Rogers J, Vu K, Haney J, Kennedy TC, Hirsch FR, Miller Y, Franklin WA, Herman JG, Baylin SB, Bunn PA, Byers T. Promoter hypermethylation of multiple genes in sputum precedes lung cancer incidence in a high-risk cohort. Cancer Res. 2006;66:3338–3344. doi: 10.1158/0008-5472.CAN-05-3408. [DOI] [PubMed] [Google Scholar]
- 83.Goessl C, Krause H, Muller M, Heicappell R, Schrader M, Sachsinger J, Miller K. Fluorescent methylation-specific polymerase chain reaction for DNA-based detection of prostate cancer in bodily fluids. Cancer Res. 2000;60:5941–5945. [PubMed] [Google Scholar]
- 84.Gonzalgo ML, Pavlovich CP, Lee SM, Nelson WG. Prostate cancer detection by GSTP1 methylation analysis of postbiopsy urine specimens. Clin Cancer Res. 2003;9:2673–2677. [PubMed] [Google Scholar]
- 85.Yegnasubramanian S, Lin X, Haffner MC, DeMarzo AM, Nelson WG. Combination of methylated-DNA precipitation and methylation-sensitive restriction enzymes (COMPARE-MS) for the rapid, sensitive and quantitative detection of DNA methylation. Nucleic Acids Res. 2006;34:e19. doi: 10.1093/nar/gnj022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Herman JG, Graff JR, Myohanen S, Nelkin BD, Baylin SB. Methylation-specific PCR: a novel PCR assay for methylation status of CpG islands. Proc Natl Acad Sci U S A. 1996;93:9821–9826. doi: 10.1073/pnas.93.18.9821. °This paper introduced the now widely used MSP analysis of methylation status of a gene.
- 87.Liu ZJ, Maekawa M. Polymerase chain reaction-based methods of DNA methylation analysis. Anal Biochem. 2003;317:259–265. doi: 10.1016/s0003-2697(03)00169-6. [DOI] [PubMed] [Google Scholar]
- 88.Licchesi JD, Herman JG. Methylation-specific PCR. Methods Mol Biol. 2009;507:305–323. doi: 10.1007/978-1-59745-522-0_22. [DOI] [PubMed] [Google Scholar]
- 89.Palmisano WA, Divine KK, Saccomanno G, Gilliland FD, Baylin SB, Herman JG, Belinsky SA. Predicting lung cancer by detecting aberrant promoter methylation in sputum. Cancer Res. 2000;60:5954–5958. [PubMed] [Google Scholar]
- 90.House MG, Guo M, Iacobuzio-Donahue C, Herman JG. Molecular progression of promoter methylation in intraductal papillary mucinous neoplasms (IPMN) of the pancreas. Carcinogenesis. 2003;24:193–198. doi: 10.1093/carcin/24.2.193. [DOI] [PubMed] [Google Scholar]
- 91.van Engeland M, Weijenberg MP, Roemen GM, Brink M, de Bruine AP, Goldbohm RA, van den Brandt PA, Baylin SB, de Goeij AF, Herman JG. Effects of dietary folate and alcohol intake on promoter methylation in sporadic colorectal cancer: the Netherlands cohort study on diet and cancer. Cancer Res. 2003;63:3133–3137. [PubMed] [Google Scholar]
- 92.Hegi ME, Diserens AC, Gorlia T, Hamou MF, de Tribolet N, Weller M, Kros JM, Hainfellner JA, Mason W, Mariani L, Bromberg JE, Hau P, Mirimanoff RO, Cairncross JG, Janzer RC, Stupp R. MGMT gene silencing and benefit from temozolomide in glioblastoma. N Engl J Med. 2005;352:997–1003. doi: 10.1056/NEJMoa043331. [DOI] [PubMed] [Google Scholar]
- 93.Goessl C, Muller M, Heicappell R, Krause H, Straub B, Schrader M, Miller K. DNA-based detection of prostate cancer in urine after prostatic massage. Urology. 2001;58:335–338. doi: 10.1016/s0090-4295(01)01268-7. [DOI] [PubMed] [Google Scholar]
- 94.Cairns P, Esteller M, Herman JG, Schoenberg M, Jeronimo C, Sanchez-Cespedes M, Chow NH, Grasso M, Wu L, Westra WB, Sidransky D. Molecular detection of prostate cancer in urine by GSTP1 hypermethylation. Clin Cancer Res. 2001;7:2727–2730. °°This exploratory study was the first demonstration of the feasibility of methylation-based detection of cancer in urine.
- 95.Roupret M, Hupertan V, Yates DR, Catto JW, Rehman I, Meuth M, Ricci S, Lacave R, Cancel-Tassin G, de la Taille A, Rozet F, Cathelineau X, Vallancien G, Hamdy FC, Cussenot O. Molecular detection of localized prostate cancer using quantitative methylation-specific PCR on urinary cells obtained following prostate massage. Clin Cancer Res. 2007;13:1720–1725. doi: 10.1158/1078-0432.CCR-06-2467. [DOI] [PubMed] [Google Scholar]
- 96.Woodson K, O’Reilly KJ, Hanson JC, Nelson D, Walk EL, Tangrea JA. The usefulness of the detection of GSTP1 methylation in urine as a biomarker in the diagnosis of prostate cancer. J Urol. 2008;179:508–511. doi: 10.1016/j.juro.2007.09.073. discussion 511-502. [DOI] [PubMed] [Google Scholar]
- 97.Vener T, Derecho C, Baden J, Wang H, Rajpurohit Y, Skelton J, Mehrotra J, Varde S, Chowdary D, Stallings W, Leibovich B, Robin H, Pelzer A, Schafer G, Auprich M, Mannweiler S, Amersdorfer P, Mazumder A. Development of a multiplexed urine assay for prostate cancer diagnosis. Clin Chem. 2008;54:874–882. doi: 10.1373/clinchem.2007.094912. [DOI] [PubMed] [Google Scholar]
- 98.Sunami E, Shinozaki M, Higano CS, Wollman R, Dorff TB, Tucker SJ, Martinez SR, Singer FR, Hoon DS. Multimarker Circulating DNA Assay for Assessing Blood of Prostate Cancer Patients. Clin Chem. 2009 doi: 10.1373/clinchem.2008.108498. [DOI] [PubMed] [Google Scholar]
- 99.Reibenwein J, Pils D, Horak P, Tomicek B, Goldner G, Worel N, Elandt K, Krainer M. Promoter hypermethylation of GSTP1, AR, and 14-3-3sigma in serum of prostate cancer patients and its clinical relevance. Prostate. 2007;67:427–432. doi: 10.1002/pros.20533. [DOI] [PubMed] [Google Scholar]
- 100.Bastian PJ, Palapattu GS, Yegnasubramanian S, Rogers CG, Lin X, Mangold LA, Trock B, Eisenberger MA, Partin AW, Nelson WG. CpG island hypermethylation profile in the serum of men with clinically localized and hormone refractory metastatic prostate cancer. J Urol. 2008;179:529–534. doi: 10.1016/j.juro.2007.09.038. discussion 534-525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Ellinger J, Haan K, Heukamp LC, Kahl P, Buttner R, Muller SC, von Ruecker A, Bastian PJ. CpG island hypermethylation in cell-free serum DNA identifies patients with localized prostate cancer. Prostate. 2008;68:42–49. doi: 10.1002/pros.20651. [DOI] [PubMed] [Google Scholar]
- 102.Cottrell S, Jung K, Kristiansen G, Eltze E, Semjonow A, Ittmann M, Hartmann A, Stamey T, Haefliger C, Weiss G. Discovery and validation of 3 novel DNA methylation markers of prostate cancer prognosis. J Urol. 2007;177:1753–1758. doi: 10.1016/j.juro.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 103.Bastian PJ, Ellinger J, Heukamp LC, Kahl P, Muller SC, von Rucker A. Prognostic value of CpG island hypermethylation at PTGS2, RAR-beta, EDNRB, and other gene loci in patients undergoing radical prostatectomy. Eur Urol. 2007;51:665–674. doi: 10.1016/j.eururo.2006.08.008. discussion 674. [DOI] [PubMed] [Google Scholar]
- 104.Suzuki M, Shigematsu H, Shames DS, Sunaga N, Takahashi T, Shivapurkar N, Iizasa T, Frenkel EP, Minna JD, Fujisawa T, Gazdar AF. DNA methylation-associated inactivation of TGFbeta-related genes DRM/Gremlin, RUNX3, and HPP1 in human cancers. Br J Cancer. 2005;93:1029–1037. doi: 10.1038/sj.bjc.6602837. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Bastian PJ, Palapattu GS, Lin X, Yegnasubramanian S, Mangold LA, Trock B, Eisenberger MA, Partin AW, Nelson WG. Preoperative serum DNA GSTP1 CpG island hypermethylation and the risk of early prostate-specific antigen recurrence following radical prostatectomy. Clin Cancer Res. 2005;11:4037–4043. doi: 10.1158/1078-0432.CCR-04-2446. [DOI] [PubMed] [Google Scholar]
- 106.Brena RM, Huang TH, Plass C. Quantitative assessment of DNA methylation: Potential applications for disease diagnosis, classification, and prognosis in clinical settings. J Mol Med. 2006;84:365–377. doi: 10.1007/s00109-005-0034-0. [DOI] [PubMed] [Google Scholar]
- 107.Zeschnigk M, Bohringer S, Price EA, Onadim Z, Masshofer L, Lohmann DR. A novel real-time PCR assay for quantitative analysis of methylated alleles (QAMA): analysis of the retinoblastoma locus. Nucleic Acids Res. 2004;32:e125. doi: 10.1093/nar/gnh122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lo YM, Wong IH, Zhang J, Tein MS, Ng MH, Hjelm NM. Quantitative analysis of aberrant p16 methylation using real-time quantitative methylation-specific polymerase chain reaction. Cancer Res. 1999;59:3899–3903. [PubMed] [Google Scholar]
- 109.McCullagh P. a. N., J. A. Generalized Linear Models. Chapman and Hall; London: 1989. [Google Scholar]
- 110.Kuppuswamy MN, Hoffmann JW, Kasper CK, Spitzer SG, Groce SL, Bajaj SP. Single nucleotide primer extension to detect genetic diseases: experimental application to hemophilia B (factor IX) and cystic fibrosis genes. Proc Natl Acad Sci U S A. 1991;88:1143–1147. doi: 10.1073/pnas.88.4.1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Singer-Sam J, LeBon JM, Dai A, Riggs AD. A sensitive, quantitative assay for measurement of allele-specific transcripts differing by a single nucleotide. PCR Methods Appl. 1992;1:160–163. doi: 10.1101/gr.1.3.160. [DOI] [PubMed] [Google Scholar]
- 112.Szabo PE, Mann JR. Allele-specific expression and total expression levels of imprinted genes during early mouse development: implications for imprinting mechanisms. Genes Dev. 1995;9:3097–3108. doi: 10.1101/gad.9.24.3097. [DOI] [PubMed] [Google Scholar]
- 113.Greenwood AD, Burke DT. Single nucleotide primer extension: quantitative range, variability, and multiplex analysis. Genome Res. 1996;6:336–348. doi: 10.1101/gr.6.4.336. [DOI] [PubMed] [Google Scholar]
- 114.Gonzalgo ML, Jones PA. Rapid quantitation of methylation differences at specific sites using methylation-sensitive single nucleotide primer extension (Ms-SNuPE) Nucleic Acids Res. 1997;25:2529–2531. doi: 10.1093/nar/25.12.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Cottrell SE, Distler J, Goodman NS, Mooney SH, Kluth A, Olek A, Schwope I, Tetzner R, Ziebarth H, Berlin K. A real-time PCR assay for DNA-methylation using methylation-specific blockers. Nucleic Acids Res. 2004;32:e10. doi: 10.1093/nar/gnh008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Jahr S, Hentze H, Englisch S, Hardt D, Fackelmayer FO, Hesch RD, Knippers R. DNA fragments in the blood plasma of cancer patients: quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001;61:1659–1665. [PubMed] [Google Scholar]
- 117.Sozzi G, Conte D, Mariani L, Lo Vullo S, Roz L, Lombardo C, Pierotti MA, Tavecchio L. Analysis of circulating tumor DNA in plasma at diagnosis and during follow-up of lung cancer patients. Cancer Res. 2001;61:4675–4678. [PubMed] [Google Scholar]
- 118.Ronaghi M, Karamohamed S, Pettersson B, Uhlen M, Nyren P. Real-time DNA sequencing using detection of pyrophosphate release. Anal Biochem. 1996;242:84–89. doi: 10.1006/abio.1996.0432. [DOI] [PubMed] [Google Scholar]
- 119.Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363, 365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- 120.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 121.Leon SA, Shapiro B, Sklaroff DM, Yaros MJ. Free DNA in the serum of cancer patients and the effect of therapy. Cancer Res. 1977;37:646–650. °The first study showing presence of DNA in the serum
- 122.Chang HW, Lee SM, Goodman SN, Singer G, Cho SK, Sokoll LJ, Montz FJ, Roden R, Zhang Z, Chan DW, Kurman RJ, Shih Ie M. Assessment of plasma DNA levels, allelic imbalance, and CA 125 as diagnostic tests for cancer. J Natl Cancer Inst. 2002;94:1697–1703. doi: 10.1093/jnci/94.22.1697. [DOI] [PubMed] [Google Scholar]
- 123.Wang BG, Huang HY, Chen YC, Bristow RE, Kassauei K, Cheng CC, Roden R, Sokoll LJ, Chan DW, Shih Ie M. Increased plasma DNA integrity in cancer patients. Cancer Res. 2003;63:3966–3968. [PubMed] [Google Scholar]
- 124.Bastian PJ, Yegnasubramanian S, Palapattu GS, Rogers CG, Lin X, De Marzo AM, Nelson WG. Molecular biomarker in prostate cancer: the role of CpG island hypermethylation. Eur Urol. 2004;46:698–708. doi: 10.1016/j.eururo.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 125.Nakayama M, Gonzalgo ML, Yegnasubramanian S, Lin X, De Marzo AM, Nelson WG. GSTP1 CpG island hypermethylation as a molecular biomarker for prostate cancer. J Cell Biochem. 2004;91:540–552. doi: 10.1002/jcb.10740. [DOI] [PubMed] [Google Scholar]
- 126.Chuang CK, Chu DC, Tzou RD, Liou SI, Chia JH, Sun CF. Hypermethylation of the CpG islands in the promoter region flanking GSTP1 gene is a potential plasma DNA biomarker for detecting prostate carcinoma. Cancer Detect Prev. 2007;31:59–63. doi: 10.1016/j.cdp.2006.11.001. [DOI] [PubMed] [Google Scholar]
- 127.Rogers CG, Gonzalgo ML, Yan G, Bastian PJ, Chan DY, Nelson WG, Pavlovich CP. High concordance of gene methylation in post-digital rectal examination and post-biopsy urine samples for prostate cancer detection. J Urol. 2006;176:2280–2284. doi: 10.1016/j.juro.2006.07.047. [DOI] [PubMed] [Google Scholar]
- 128.Tomlins SA, Rubin MA, Chinnaiyan AM. Integrative biology of prostate cancer progression. Annu Rev Pathol. 2006;1:243–271. doi: 10.1146/annurev.pathol.1.110304.100047. [DOI] [PubMed] [Google Scholar]
- 129.Bryzgunova OE, Morozkin ES, Yarmoschuk SV, Vlassov VV, Laktionov PP. Methylation-specific sequencing of GSTP1 gene promoter in circulating/extracellular DNA from blood and urine of healthy donors and prostate cancer patients. Ann N Y Acad Sci. 2008;1137:222–225. doi: 10.1196/annals.1448.039. [DOI] [PubMed] [Google Scholar]
- 130.Jemal A, Tiwari RC, Murray T, Ghafoor A, Samuels A, Ward E, Feuer EJ, Thun MJ. Cancer statistics, 2004. CA Cancer J Clin. 2004;54:8–29. doi: 10.3322/canjclin.54.1.8. [DOI] [PubMed] [Google Scholar]
- 131.Sylvester JE, Blasko JC, Grimm PD, Meier R, Malmgren JA. Ten-year biochemical relapse-free survival after external beam radiation and brachytherapy for localized prostate cancer: the Seattle experience. Int J Radiat Oncol Biol Phys. 2003;57:944–952. doi: 10.1016/s0360-3016(03)00739-9. [DOI] [PubMed] [Google Scholar]
- 132.Kupelian PA, Elshaikh M, Reddy CA, Zippe C, Klein EA. Comparison of the efficacy of local therapies for localized prostate cancer in the prostate-specific antigen era: a large single-institution experience with radical prostatectomy and external-beam radiotherapy. J Clin Oncol. 2002;20:3376–3385. doi: 10.1200/JCO.2002.01.150. [DOI] [PubMed] [Google Scholar]
- 133.Kuban DA, Thames HD, Levy LB, Horwitz EM, Kupelian PA, Martinez AA, Michalski JM, Pisansky TM, Sandler HM, Shipley WU, Zelefsky MJ, Zietman AL. Long-term multi-institutional analysis of stage T1-T2 prostate cancer treated with radiotherapy in the PSA era. Int J Radiat Oncol Biol Phys. 2003;57:915–928. doi: 10.1016/s0360-3016(03)00632-1. [DOI] [PubMed] [Google Scholar]
- 134.Karakiewicz PI, Shariat SF, Palapattu GS, Gilad AE, Lotan Y, Rogers CG, Vazina A, Gupta A, Bastian PJ, Perrotte P, Sagalowsky AI, Schoenberg M, Lerner SP. Nomogram for predicting disease recurrence after radical cystectomy for transitional cell carcinoma of the bladder. J Urol. 2006;176:1354–1361. doi: 10.1016/j.juro.2006.06.025. discussion 1361-1352. [DOI] [PubMed] [Google Scholar]
- 135.Kattan MW, Wheeler TM, Scardino PT. Postoperative nomogram for disease recurrence after radical prostatectomy for prostate cancer. J Clin Oncol. 1999;17:1499–1507. doi: 10.1200/JCO.1999.17.5.1499. [DOI] [PubMed] [Google Scholar]
- 136.Roupret M, Hupertan V, Catto JW, Yates DR, Rehman I, Proctor LM, Phillips J, Meuth M, Cussenot O, Hamdy FC. Promoter hypermethylation in circulating blood cells identifies prostate cancer progression. Int J Cancer. 2008;122:952–956. doi: 10.1002/ijc.23196. [DOI] [PubMed] [Google Scholar]