Abstract
Objectives
This study tested the hypothesis that the fecal bacterial genera of breast-fed (BF) and formula-fed (FF) infants differ and that human milk oligosaccharides (HMO) modulate the microbiota of BF infants.
Methods
Fecal samples were obtained from BF (n = 16) or FF (n = 6) infants at 3-month postpartum. Human milk were collected on the same day when feces were collected. The microbiota was assessed by pyrosequencing of bacterial 16S rRNA genes. HMO were measured by HPLC-Chip time-of-flight mass spectrometry.
Results
The overall microbiota of BF differed from that of FF (P = 0.005). Compared to FF, BF had higher relative abundances of Bacteroides, lower proportions of Clostridium XVIII, Lachnospiracea incertae sedis, Streptococcus, Enterococcus and Veillonella (P < 0.05). Bifidobacterium predominated in both BF and FF infants, with no difference in abundance between the two groups. The most abundant HMO were lacto-N-tetraose + lacto-N-neotetraose (LNT + LNnT, 22.6%), followed by 2′-fucosyllactose (2′FL, 14.5%) and lacto-N-fucopentaose I (LNFP I, 9.5%). Partial least squares regression of HMO and microbiota showed several infant fecal bacterial genera could be predicted by their mothers’ HMO profiles and the important HMO for the prediction of bacterial genera were identified by variable importance in the projection scores.
Conclusions
These results strengthen the established relationship between HMO and the infant microbiota, identify statistical means whereby infant bacterial genera can be predicted by milk HMO. Future studies are needed to validate these findings and determine if supplementation of formula with defined HMO could selectively modify the gut microbiota.
Keywords: gut microbiota, infants, human milk oligosaccharide, breastfed
INTRODUCTION
The development of the intestinal microbiota occurs primarily during infancy. Mutualistic interactions between the colonizing intestinal bacteria and the host are essential for healthy intestinal and immunological development (1). The stepwise microbial colonization process appears to have a long-lasting influence on the risk of not only gastrointestinal disease, but also allergic, autoimmune and metabolic diseases, in later life (2,3). Early microbial programming begins in utero and is substantially modulated by host and environmental factors, including duration of gestation, mode of delivery, environmental microbes, antibiotic use and diet (4,5).
Human milk is the optimal diet for infants and exclusive breastfeeding is recommended for the first 6-mos of life; however, a large proportion of U.S. babies (~81%) are no longer exclusively breastfed by 6 months-of-age (6). The influence of feeding type, breast or formula, on the composition of the microbiota is currently equivocal. Many studies have shown that the microbiota of breast-fed (BF) infants is predominated by Bifidobacterium, while formula-fed (FF) infants are often colonized by more diverse microbiota, with a lower abundance of Bifidobacterium (7). In contrast, other studies reported that all infants were colonized by Bifidobacterium, with no differences in prevalence or abundance between BF and FF infants (8,9).
Emerging evidence support a role for human milk oligosaccharides (HMO) in shaping the composition of the infant gut microbiota. HMO are the third most abundant component of human milk after lactose and lipid, present at high concentrations (~20 g/L in colostrum; 5–10 g/L in mature milk) (10). Additionally, HMO exhibit great structural diversity, with more than 200 different structures being defined (11). HMO are resistant to enzymatic hydrolysis in the upper gastrointestinal tract and the majority of HMO (> 90%) reach the colon (12), where they serve as the primary substrate for growth of specific subsets of bacteria in the intestine of BF infants (13,14). Consumption of HMO by gut bacteria has been studied in vitro by measuring the growth of pure bacterial isolates in culture media containing individual or mixtures of HMO (14). Others have investigated fermentation of individual HMO or a mixture of HMO ex vivo using fecal/intestinal microbiota from infants or piglets (15, 16). Tracking HMO excretion can provide insight into the selective utilization by gut microbes in human infants in vivo (17, 18). However, little is known about the association between composition of the gut microbiota and HMO profiles in vivo. Therefore, the goal of this study was to compare the fecal microbial composition between BF and FF infants and to examine the ability of milk HMO to predict bacterial genera in BF infants.
MATERIALS AND METHODS
Study subjects and design
Healthy, full term, vaginally-delivered, exclusively BF (n=16) or FF (n=6) (Enfamil LIPIL, Mead Johnson Nutrition, Evansville, IN) infants were eligible for enrollment into the study. Details of the subject recruitment, inclusion and exclusion criteria have been previously described (19). Both mothers and infants were medically certified as healthy (asymptomatic and with no clinical indication of disease) during the study and the mothers consumed their normal diet. Enrolled infants who subsequently received antibiotic treatment were excluded from the study. Briefly, mothers of infants were recruited into the study between the third trimester pregnancy and 1 mo postpartum. We focused on recruiting second parity mothers who had either exclusively breastfed or formula-fed their first infant to increase our likelihood of enrolling mothers who were secure in their child feeding decision. Enfamil LIPIL formula were provided to mother who decided formula feed their infants prior delivery so that the infant began on the formula immediately after birth. All infants were fed ad libitum. Freshly-voided stool samples were collected from the infants’ diaper by the parent at 3 months-of-age using a sterile spoon. Samples were placed into sterile 2 mL tubes (Corning Incorporated, Corning, NY). Human milk was collected on the same day that fecal samples were collected. Milk was collected by expressing the contents of one breast, while the infant nursed on the other breast. To ensure the ‘full’ content of the breast had been expressed, participants continued to express the mammary gland until milk flow had subsided. The milk sample was mixed in order to obtain a homogenous sample, from which 30 mL was placed into a sterile 50 mL conical tube (Corning Incorporated, Corning, NY) and remaining milk was retained by the parent. All stool and milk samples were stored at 4°C for 2–3h, before being transported on ice to the laboratory, where they were stored at −80°C. All study procedures were approved by the University of Illinois Institutional Review Board and informed consent was obtained from parents prior to participation in the study.
HMO analysis
HMO were extracted, reduced and purified from milk samples according to previously described methods (20,21). HMO composition was profiled using an Agilent 6210 high performance liquid chromatography-chip time-of-flight mass spectrometry (HPLC-Chip/TOF MS) system equipped with both a capillary pump for sample loading and a nanopump for sample separation (Agilent Technologies, Santa Clara, CA) as previously described (20,21). Data were collected in the positive mode and calibrated by a dual nebulizer electrospray source with a wide range of internal calibrant ions: m/z 118.086, 322.048, 622.029, 922.010, 1221.991, 1521.972, 1821.952, 2121.933, 2421.914, and 2721.895. HMO identification and quantitation was performed using Agilent Mass Hunter Qualitative Analysis software (version B.03.01) as described by Totten et al. (22). The relative amount of each oligosaccharide species was calculated by normalizing the absolute abundance of the individual species to the total oligosaccharide ion abundance in each sample, yielding a relative abundance expressed as a percentage of the total.
DNA isolation from fecal samples
DNA was extracted using a modification of the method of Yu and Morrison (23). The detailed protocol has been previously described (9). DNA quality was checked on a 1% agarose gel following ethidium bromide staining. DNA from 3 to 4 extractions per sample were pooled and concentration quantified on a NanoDrop 1000 spectrophotometer (NanoDrop Technologies, Wilmington DE).
Analysis of fecal microbiota by pyrosequencing of 16S rRNA genes
Amplification of the V1–V3 regions of the bacterial 16S rRNA genes was performed with fusion primers. Each forward primer (from 5′ to 3′) included: GS FLX Titanium Primer A (CCATCTCATCCCTGCGTGTCTCCGACTCAG), a Multiplex Identifier that was unique to each sample, and 27F-DegS (24). The reverse primer (from 5′ to 3′) contained Primer B (CCTATCCCCTGTGTGCCTTGGCAGTCTCAG) and 534R (25). The FastStart High Fidelity PCR System, dNTPack (Roche Applied Science, Indianapolis, IN) was used for PCR amplification. The PCR reaction mixture contained 0.2 μM of each primer, 10 ng of template DNA, 5 μl of 10 × PCR reaction buffer, 200 μM of each deoxyribonucleotide triphosphate, 2.5 μL bovine serum albumin (New England Biolabs, Ipswich, MA) at 1 mg/mL (final concentration 100 μg/mL), 1.8 mM MgCl2 and 1.25 U of FastStart Hi-Fi enzyme blend in a total volume of 25 μL. PCR was performed in a DNAEngine (Bio-Rad, Hercules, CA) under the following conditions: 94C for 3 min followed by 25 cycles of 94°C for 30 sec, 56°C for 30 sec, and 72°C for 1 min, and a final elongation step at 72°C for 7 min. After PCR, the amplicons from 3 separate reactions were pooled and purified using Agencourt AMPure XP according to manufacturer instructions (Beckman Coulter, Inc., Brea, CA). Prior to pyrosequencing, DNA concentration was measured with Quant-iT PicoGreen dsDNA Assay Kits (Life technologies, Grand Island, NY) and DNA quality was assessed using a 2100 Bioanalyzer (Agilent, Santa Clara, CA). The amplicons were mixed in equimolar concentration and sequenced at the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois using 454 Life Sciences Genome Sequencer FLX with GS FLX Titanium series reagents (Roche Applied Science, Indianapolis, IN).
Sequence processing
The 16S rDNA sequences were processed and analyzed using the QIIME pipeline (v1.6.0 (26). Sequences were removed from further analysis if their length was outside the range of 400–600 nt, or if they contained ambiguous bases, primer mismatches, homopolymer run greater than six nucleotides, or uncorrectable barcodes. The remaining sequences were denoised with the Denoiser algorithm within QIIME and clustered into operational taxonomic units (OTUs) at 97% pairwise identity using the UCLUST algorithm within QIIME. The representative sequences from each OTU were picked and the chimera sequences were identified via Chimera Slayer. After removal of chimeras, the remaining sequences were aligned to the Greengenes imputed core reference alignment (27) using PyNAST and the alignment were filtered to remove highly variable regions and columns comprised of only gaps using a lane mask. The phylogenetic tree was constructed from filtered alignment using FastTree (28) and unweighted UnFrac distance matrix (29) was generated from phylogenetic tree. The representative sequence of each OTU was assigned to different taxonomic levels using Ribosomal Database Project (RDP release 11.1) naïve Bayesian rRNA Classifier at 80% confidence level (30). Alpha-diversity (observed OTUs, Chao1 and ACE estimators, Shannon and Simpson reciprocal indices) was calculated using QIIME after rarefying to an equal number of reads (7,800) for all samples to control for unequal sampling effort.
Statistical analysis
To detect whether the structure of the bacterial communities between BF and FF infants differed, principal co-ordinate analysis (PCoA) and distance-based redundancy analysis (dbRDA) were performed on unweighted UniFrac distance using QIIME and the capscale command of vegan package of R, respectively (31).
Univariate statistical analysis was performed using PROC MIXED procedure of SAS version 9.2 (SAS Institute, Cary, NC). When the data were not normally distributed, the Mann-Whitney test was used. Spearman’s rank correlation test was applied to explore relationship between bacterial genera. Statistical significance was set at P < 0.05.
The associations between HMO profiles and each bacterial genus were modeled by partial least squares (PLS) regression. The HMO which contributed most to the relationship were identified by calculating variable importance in the projection (VIP) scores (32). Data were log transformed and mean centered before the PLS regression. The predictive performance of PLS model was evaluated by tenfold cross-validation. A variable with VIP ≥ 1.2 was considered influential. The direction of correlation (positive or negative) was determined according to PLS regression coefficients (Beta). Spearman correlation, PLS regression and VIP analyses were performed under MATLAB R2011b environment (The Mathworks, Natick, MA).
RESULTS
Demographics and growth of subjects
A total of 16 BF and 6 FF infants were recruited for the study and most of the infants were Caucasian (Table 1). There was no difference in mean age between mothers of BF and FF infants. The sex distribution and birth length of infants were similar in both groups. Body weight at birth and 3 mos of age did not differ between BF and FF infants (Table 1).
TABLE 1.
Demographics and growth of subjects
| BF, n = 16 | FF, n = 6 | |
|---|---|---|
| Maternal age, yr* | 29.1 ± 4.7 | 30.7 ± 2.4 |
| Infant sex | 4 female, 12 male | 2 female, 4 male |
| Infant ethnicity | 13 Caucasian 2 African/Caucasian 1 African American |
6 Caucasian |
| Length at birth, cm* | 53.5 ± 2.9 | 51.0 ± 2.5 |
| Body weight, kg* | ||
| Birth | 3.73 ± 0.55 | 3.50 ± 0.20 |
| 3 mo | 6.70 ± 0.58 | 6.45 ± 0.96 |
Mean ± SEM.
BF, breast-fed; FF, formula-fed.
HMO composition
In total, 141 types of oligosaccharides were detected and the average numbers of HMO in all milk samples was 63.4 ± 1.4. Nonfucosylated neutral oligosaccharides accounted for 25.4 ± 2.26% of total HMO. Fucosylated, sialylated or both fucosylated and sialylated oligosaccharides comprised 61.1± 2.46, 11.0 ± 1.65 and 2.53 ± 0.41%, respectively (Table 2). The predominant HMO are shown in Table 2. Lacto-N-tetraose and lacto-N-neotetraose (LNT + LNnT; 22.6%) together were the most predominant HMO, followed by 2′-fucosyllactose (2′FL; 14.5%), lacto-N-fucopentaose I (LNFP I; 9.48%), lacto-N-fucopentaoseII (LNFP II; 8.17%) and lactodifucotetraose (LDFT; 6.61%). Other HMO accounted for less than 5% of total HMO.
TABLE 2.
Composition of human milk oligosaccharides in breast milk
| Name | % of total HMO* | Name | % of total HMO* |
|---|---|---|---|
| Nonfucosylated neutral | 25.4 ± 2.26 | FS-LNnH I | 1.27 ± 0.91 |
| Fucosylated | 61.1 ± 2.46 | m/z 855.3 @ 18 min | 1.15 ± 0.65 |
| Sialylated | 11.0 ± 1.65 | LNDFH II | 1.09 ± 1.07 |
| Fucosylated & sialylated | 2.53 ± 0.41 | m/z 636.3 @ 16 min | 0.95 ± 0.78 |
|
|
|||
| LNT+LNnT | 22.6 ± 7.36 | 3′SL | 0.92 ± 0.47 |
| 2′FL | 14.5 ± 12.1 | m/z 1074.4 @ 21 min | 0.88 ± 1.03 |
| LNFP I | 9.48 ± 5.62 | 3′FL | 0.87 ± 0.69 |
| LNFP II | 8.17 ± 4.33 | m/z 1439.5@ 23 min | 0.85 ± 0.36 |
| LDFT | 6.61 ± 3.66 | LSTc | 0.76 ± 0.14 |
| MFLNH III | 3.82 ± 3.38 | IFLNH III | 0.70 ± 1.01 |
| LSTb | 3.14 ± 3.48 | m/z 709.3 @ 12 min | 0.69 ± 0.34 |
| DFLNHa | 2.47 ± 1.90 | MSLNnH | 0.67 ± 0.55 |
| LNnH | 2.38 ± 1.60 | LNDFH I | 0.58 ± 0.19 |
| DSLNT | 1.95 ± 2.37 | 5130a | 0.54 ± 0.33 |
| LNH | 1.93 ± 1.24 | m/z 490.2 @ 14 min | 0.53 ± 0.73 |
| MFpLNH IV | 1.91 ± 1.24 | DFpLNH II | 0.51 ± 0.42 |
| DFLNHb | 1.89 ± 2.30 | ||
Mean ± SEM, n = 16.
Only HMO with relative abundance > 0.5% are shown.
2′FL, 2′-fucosyllactose; 3′FL, 3′-fucosyllactose; 3′SL, 3′-sialyllactose; DFLNHa, difucosyllacto-N-hexaose a; DFLNHb, difucosyllacto-N-hexaose b; DFpLNH II, difucosyl-para-lacto-N-hexaose II; DSLNT, disialyllacto-N-tetraose; FS-LNnH I, fucosyl-sialyl-lacto-N-neohexaose I; HMO, human milk oligosaccharides; IFLNH III, isomeric fucosylated lacto-N-hexaose III; LDFT, lactodifucotetraose; LNDFH I, lacto-N-difucohexaose I; LNDFH II, lacto-N-difucohexaose II; LNFP I, lacto-N-fucopentaose I; LNFP II, lacto-N-fucopentaose II; LNH, lacto-N-hexaose; LNnH, lacto-N-neohexaose; LNT + LNnT, lacto-N-tetraose + lacto-N-neotetraose; LSTb, sialyllacto-N-tetraose b; LSTc, sialyllacto-N-tetraose c; MFLNH III, monofucosyllacto-N-hexaose III; MFpLNH IV, monofucosyl-para-lacto-N-hexaose IV; MSLNnH: monosialyllacto-N-neohexaose I
Fecal microbiota of BF and FF infants by pyrosequencing
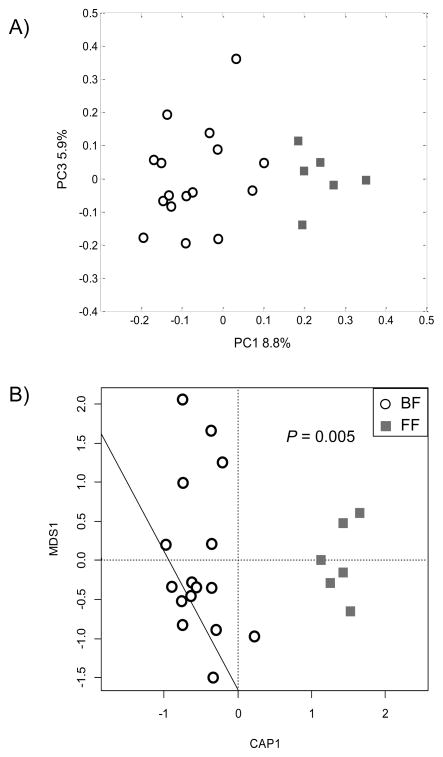
Pyrosequencing of the V1–V3 regions of the 16S rRNA gene amplicons yielded 417,344 total reads with an average read length of 453 bp. After performing the quality control depletions as above, 321,822 sequences with a mean of 10,734 sequences (range = 7,863–13,410) per sample were utilized for further analysis. Unweighted UniFrac PCoA revealed that the fecal microbial structure of BF infants differed from that of FF infants (Figure 1A). This was confirmed by dbRDA of unweighted UniFrac distances (P = 0.005; Figure 1B).
Figure 1.
PCoA (A) and dbRDA (B) based on unweighted UniFrac distances generated from fecal samples of 3 mos-old BF and FF infants. n = 16 (BF), n = 6 (FF). BF, breast-fed; dbRDA, distance-based redundancy analysis; FF, formula-fed; PCoA, principal co-ordinate analysis.
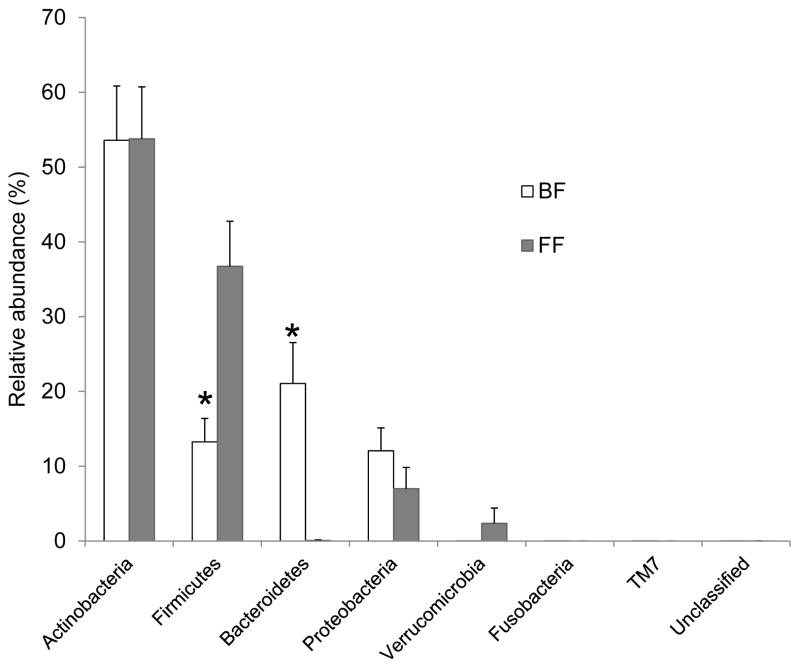
To identify which bacteria differed between BF and FF infants, the sequences were classified against RDP Classifier (Version 2.6 trained on 16S rRNA training set 9). In total, 7 phyla and 62 genera were identified. Actinobacteria was the most abundant phylum (~53 %), with no difference between BF and FF infants (Figure 2). Other bacterial phyla were Firmicutes, Bacteroidetes, Proteobacteria, Verrucomicrobia, Fusobacteria and MT7. Breast-fed infants harbored greater relative abundance of Bacteroidetes, while FF infants had higher Firmicutes (P < 0.05; Figure 2).
Figure 2.
Relative abundances of bacterial phyla within fecal microbiota of BF and FF infants at 3 mo of age. n = 16 (BF), n = 6 (FF). *FF differed from BF infants, Mann-Whitney test, P < 0.05. BF, breast-fed; FF, formula-fed.
The relative abundances of predominating bacterial genera occurring in infant feces are shown in Table 3. Bacteroides were greater in BF than FF infants. In contrast, fecal clostridium XVIII, Lachnospiracea incertae sedis, Streptococcus, Blautia, Clostridium XI, Clostridium sensu stricto, Eubacterium, Erysipelotrichaceae incertae sedis and Haemophilus were lower in BF than FF infants (P < 0.05). Bifidobacterium represented > 50% of the total sequences in both BF and FF infants, with no difference between the two groups.
TABLE 3.
Relative abundances of bacterial genera within fecal microbiota of BF and FF infants, Median (25, 75 percentiles)
| Bacterial genus | BF, n =16 | FF, n = 6 | P value |
|---|---|---|---|
| Actinobacteria | |||
| Bifidobacterium | 59.8 (36.1, 76.9) | 54.2 (41.3, 65.3) | 0.796 |
| Rothia | 0.11 (0.01, 0.36) | 0.03 (0.01, 0.04) | 0.318 |
| Bacteroidetes | |||
| Bacteroides | 13.4 (2.35, 41.0) | 0.03 (0.01, 0.06) | 0.009 |
| Parabacteroides | 0 (0, 0.21) | 0 (0, 0) | 0.068 |
| Firmicutes | |||
| Clostridium XVIII | 0 (0, 1.39) | 4.64 (2.23, 8.40) | 0.006 |
| Lachnospiracea incertae sedis | 0 (0, 0.18) | 0.88 (0.86, 0.94) | 0.001 |
| Streptococcus | 0.34 (0.06, 1.08) | 4.87 (2.69, 5.95) | 0.001 |
| Enterococcus | 0.14 (0.01, 0.41) | 0.50 (0.09, 0.93) | 0.112 |
| Veillonella | 0.60 (0.04, 1.37) | 0.22 (0.11, 1.10) | 0.912 |
| Blautia | 0 (0, 0) | 0.01 (0, 4.04) | 0.021 |
| Flavonifractor | 0 (0, 0.05) | 0.01 (0, 0.03) | 0.432 |
| Clostridium XI | 0 (0, 0.02) | 0.23 (0.19, 0.47) | 0.003 |
| Clostridium sensu stricto | 0 (0, 0.02) | 0.56 (0.28, 0.75) | 0.004 |
| Clostridium XlVa | 0 (0, 0.07) | 0.01 (0, 0.02) | 0.932 |
| Eubacterium | 0 (0, 0) | 0.04 (0, 0.39) | 0.004 |
| Erysipelotrichaceae incertae sedis | 0 (0, 0) | 0.17 (0.04, 0.55) | 0.005 |
| Lactobacillus | 0 (0, 0.02) | 0 (0, 0) | 0.146 |
| Anaerostipes | 0 (0, 0) | 0 (0, 0.09) | 0.123 |
| Staphylococcus | 0.02 (0, 0.13) | 0 (0,0.01) | 0.056 |
| Proteobacteria | |||
| Escherichia/Shigella | 6.11 (2.13, 14.1) | 4.49 (1.49, 6.57) | 0.631 |
| Klebsiella | 0.03 (0, 0.86) | 0.27 (0.11, 0.70) | 0.279 |
| Sutterella | 0 (0, 0.18) | 0 (0, 0) | 0.581 |
| Haemophilus | 0.06 (0, 0.17) | 0 (0, 0) | 0.03 |
| Morganella | 0 (0, 0) | 0 (0.0) | 0.61 |
| Verrucomicrobia | |||
| Akkermansia | 0 (0,0) | 0 (0, 1.89) | 0.096 |
Only bacterial genera with mean relative abundance > 0.05% were analyzed.
P values were obtained by Mann–Whitney test.
BF, breast-fed; FF, formula-fed.
To compare diversity within samples, sequences were rarefied to an equal number of reads (7,800) for all samples and observed OTUs, Chao 1 and ACE estimators, Shannon and reciprocal Simpson indices were calculated (Table 4). The observed OTUs, Chao 1 and ACE estimators did not differ between BF and FF infants. Similarly, no differences in the Shannon and reciprocal Simpson indices were detected between the two groups.
TABLE 4.
Diversity measures obtained from fecal samples of BF and FF infants, mean ± SEM
| Observed OTUs, n | Shannon | Reciprocal Simpson | ACE | Chao1 | |
|---|---|---|---|---|---|
| BF, n = 16 | 48.0 ± 2.4 | 2.75 ± 0.11 | 4.29 ± 0.35 | 83.4 ± 5.37 | 92.7 ± 10.6 |
| FF, n = 6 | 60.0 ± 3.3 | 3.10 ± 0.23 | 4.83 ± 0.73 | 87.8 ± 7.57 | 89.9 ± 7.03 |
| P value | 0.0894 | 0.1299 | 0.4645 | 0.6605 | 0.855 |
All samples were rarefied to 7,800 sequences.
P values were obtained by one-way ANOVA.
ACE, abundance-base coverage estimator; BF, breast-fed; FF, formula-fed; OTU, operational taxonomic unit.
Relationship between bacterial genera
In order to assess the relationship between the members of fecal microbiota, Spearman correlation analyses of relative abundances of bacterial genera were performed. In BF infants, the relative abundance of Enterococcus was positively correlated with abundances of Bifidobacterium, Streptococcus and Veillonella, while negatively correlated with Bacteroides and Clostridium XVIII (P < 0.05; Table 5). Significant negative correlations were also detected between Bifidobacterium and Bacteroides, Escherichia/Shigella and Klebsiella in BF infants. The only significant correlations found in FF infants were negative correlations between Bifidobacterium and Clostridium XVIII and between Veillonella and Klebsiella (Table 5).
TABLE 5.
Correlation between relative abundances of bacterial genera detected in feces of BF or FF infants
| Bacterial genus | Bacterial genus | BF, n = 16 | FF, n = 6 |
|---|---|---|---|
| Bifidobacterium | Bacteroides | −0.75* | 0.14 |
| Bifidobacterium | Clostridium XVIII | −0.45 | −0.89* |
| Bifidobacterium | Enterococcus | 0.53* | −0.54 |
| Bacteroides | Enterococcus | −0.53* | 0.66 |
| Escherichia/Shigella | Klebsiella | −0.51* | 0.49 |
| Clostridium XVIII | Enterococcus | −0.51* | 0.66 |
| Streptococcus | Enterococcus | 0.68* | 0.26 |
| Enterococcus | Veillonella | 0.52* | −0.66 |
| Veillonella | Klebsiella | −0.04 | −0.89* |
Values were Spearman correlation coefficients,
P < 0.05.
Only bacterial genera present in > 50% infants and with mean relative abundances > 0.5% were analyzed.
BF, breast-fed; FF, formula-fed.
Association between HMO and bacterial genus
The associations between HMO profiles and each bacterial genus were modeled by PLS regression and the influential HMO for the prediction of bacterial genus were identified by VIP scores. PLS regression showed several bacterial genera detected in infant feces, including Bifidobacterium, Bacteroides, Enterococcus, Veillonella, and Rothia, could be predicted by their mothers’ HMO profiles. As shown in Table 6 and Supplemental Figure 1, each bacterial genus revealed an association with multiple HMO. For example, the relative abundance of fecal Bifidobacterium was positively linked with the presence of LNFP I, monofucosyllacto-N-hexaose III (MFLNH III), sialyllacto-N-tetraose b (LSTb) and disialyllacto-N-tetraose (DSLNT) and negatively linked with the presence of 2′FL and LDFT in human milk. Furthermore, most of HMO were associated with multiple bacterial genera, for example, 2′FL was positively linked to Bacteroides, but negatively linked to Bifidobacterium, Enterococcus, Veillonella, and Rothia
TABLE 6.
HMO showing influential effects for the prediction of bacterial genera in BF infants
| Bacterial genus | MSE | Influential HMO (VIP ≥ 1.2)
|
|
|---|---|---|---|
| Positive | Negative | ||
| Bifidobacterium | 0.258 | MFLNH III, LSTb, LNFP I, DSLNT | 2′FL, LDFT |
| Bacteroides | 0.686 | 2′FL, LNFP I, LDFT | LSTb, DFLNHa, DSLNT |
| Veillonella | 0.142 | LNFP I, DSLNT, LNFP II | 2′FL |
| Enterococcus | 0.068 | DSLNT | 2′FL, LDFT |
| Rothia | 0.014 | LDFT, LSTb | 2′FL, MFLNH III, DSLNT |
Data were log transformed and mean centered prior to PLS regression. The predictive performance was evaluated by ten-fold cross validation. Only bacterial genera presented in > 50% BF infants were analyzed.
The direction of correlation (positive or negative) was determined by calculating PLS regression coefficients.
2′FL, 2′-fucosyllactose; BF, Breast-fed; DFLNHa, difucosyllacto-N-hexaose a; DSLNT, disialyllacto-N-tetraose; HMO, human milk oligosaccharides; LDFT, lactodifucotetraose; LNFP I, lacto-N-fucopentaose I; LNFP II, lacto-N-fucopentaose II; LSTb, sialyllacto-N-tetraose b; MFLNH III, monofucosyllacto-N-hexaose III. MSE, mean-squared errors for PLS model; PLS, partial least square; VIP, variable importance in the projection.
DISCUSSION
Feeding mode is one of the most important determinants of gut microbial diversity in neonates; however, its impact on the composition of the infant microbiota is often contradictory (3). These inconsistencies in the results may arise from the different analytical approaches used to enumerate the microbiota, geographically distinct infant groups studied or the variability in the composition of infant formula. To reduce those variations, we applied a high-throughput sequencing approach, enrolled infants from same geographic region and fed all FF infants with same formula throughout the experimental period. In agreement with several previous studies (7, 9), our PCoA and dbRDA analysis showed that the fecal microbiota composition of BF differed from that of FF infants. BF infants harbored greater relative abundance of Bacteroides, which belongs to Bacteroidetes, while FF infants had higher abundances of bacterial genera classified as Firmicutes, such as Clostridium XVIII, Lachnospiracea incertae sedis, Streptococcus, Enterococcus and Veillonella.
Bifidobacterium constitute a significant portion of the intestinal microbiota and are frequently used as probiotics to provide health-promoting benefits on their host (33). Colonization of Bifidobacterium in the neonatal intestine tract has been extensively studied; however, the results are often contradictory. Many studies showed that that Bifidobacterium rapidly dominated the microbiota in BF infants (34); while others reported that Bifidobacterium occurred in relatively low frequency and abundance in the fecal microbiota of BF infants (8). It is now general accepted that Bifidobacterium are usually highly abundant in BF infants and that studies that deviated from this suffered from methodological or sampling errors (35). For example, forward primer used in the study of Palmer and coworkers (8) has a three base pair mismatch against B. longum, and Bifidobacterium genus in general do not have 100% sequence identity to the forward primer.
Several older studies reported that FF infants harbor a lower abundance of Bifidobacterium compared with BF infants (34), whereas a recent review, which summarized the studies performed after 1980, concluded that in most studies Bifidobacterium are found equally often and in similar counts in BF and FF infants (36). In the current study, Bifidobacterium were predominant (> 50% of sequences) in the feces of both BF and FF infants with no difference in the relative abundances between the two groups. The high abundance of Bifidobacterium detected in FF infants in the current study suggests that the formula was able to support the growth of Bifidobacterium. In recent years, several approaches have been used to improve infant formulas to induce a microbiota profile more similar to that in breast-fed infants. These approaches, including providing an optimal ratio of casein and whey protein, and adding prebiotics or probiotics to infant formula, have been successful in increasing bifidobacteria in formula-fed infants (37, 38). The formula used in this study did not contain added prebiotics, although currently marketed Enfamil does contain 4 g/L of a 1:1 mixture of polydextrose and galactooligosaccharides. In terms of protein composition, the formula was whey-predominant and studies have shown whey-predominant formula induce a fecal microbiota generally closer to that of BF babies than did a casein-predominant formula (38). Although similar proportions of Bifidobacterium genera were detected in BF and FF infants in our study, the Bifidobacterium composition at species level could differ between the two groups, as previously demonstrated. For example, Harrman and colleagues showed that B. longum subsp. infantis, B. longum subsp. longum and B. breve were the predominant species found in BF infants, while the microbiota of FF infants contained relatively more Bifidobacterium catenulatum and Bifidobacterium adolescentis, two species that are commonly found in adults (39)
Bacteroides are predominant in the gut of human adults and several studies have confirmed that Bacteroides also dominate the intestinal microbiota of some infants (40, 41). In our study, Bacteroides represented the second most predominant bacterial genus in BF infants, after Bifidobacterium, and a significantly greater proportion of Bacteroides was detected in BF compared to FF infants (13.4% vs. 0.03%). The presence of higher levels of Bacteroides may be beneficial for the BF infant, as members of Bacteroides have been shown to exert immunomodulatory proprieties on the host (42). For example, polysaccharide A (PSA) produced by Bacteroides fragilis directs the cellular and physical maturation of the host immune system, specifically promoting the functional development of CD4+ T-cells (41). In addition, Bacteroides spp. have extensive machinery to metabolize complex polysaccharides (such as starch, pectin and host-derived glycan) (43). Thus, the presence of Bacteroides in the intestine of BF infants may confer stability and adaptability to microbiota during the transition from human milk to solid foods (44). Furthermore, degradation of polysaccharides by Bacteroides produces short-chain fatty acids, which contribute significantly to host nutrition and overall health of the colon (45).
Differences in the ratio of Firmicutes-to-Bacteroidetes were detected between lean and obese mice and human adults (46). Compared to lean mice, the cecal microbiota of obese mice had 50% fewer Bacteroidetes, and correspondingly more Firmicutes (46). Human adult studies have shown that the proportion of Bacteroidetes is decreased in obese individuals by comparison with lean people, and that increases in Bacteroidetes and reductions in Firmicutes has been documented with weight loss (47). In our study, a higher Firmicutes-to-Bacteroidetes ratio was detected in FF than BF infants; however, no differences in body weight between the two groups were observed during the study period. Previous studies have shown breastfeeding decreases children’s risk of obesity (48); therefore, the impact of the higher ratio of Firmicutes-to-Bacteroidetes in FF infants on the development of overweight and obesity later in life deserves further investigation.
Similar to the findings of a recent study by Jost and coworkers (40), the proportion of Bifidobacterium was inversely correlated with the proportion of Bacteriodes; however, such relationship was detected only in BF, not in FF infants. This may be due in part to the presence of HMO in human milk. Human milk contains large quantity of structurally-diverse oligosaccharides (10), while bovine milk, the basis for most infant formula, contains only trace amounts of predominantly siallylated oligosaccharides (49). In vitro fermentation studies have shown that some species of Bifidobacterium (e.g. B. longum subsp. infantis and B. bifidus) and Bacteroides (e.g. B. thetaiotaomicron, B. vulgatus and B. fragilis) can grow efficiently in minimal medium containing HMO as the sole carbon source (50, 51). Moreover, whole genome transcriptional profiling has revealed that some members of Bifidobacterium and Bacteroides express glycoside hydrolase and intestinal membrane transporters that are essential for the degradation of HMO (51, 52). Thus, the inverse correlation between the two genera may result from competition for HMO as metabolic substrates when both Bifidobacterium and Bacteroides are present in the gut of BF infants. In addition, inter-individual differences in HMO composition, such as the structural complexity of the HMO, may be an important selective force, since previous studies have demonstrated that Bifidobacterium (e.g. B. longum subsp. infantis) preferentially consume short HMO (53), while Bacteroides have the capacity to utilize a broad range of HMO, with a slight preference for larger ones (50). For example, a short HMO, LNnT, selectively expanded the abundance of B. longum subsp. infantis relative to B. thetaiotaomicron in bi-associated gnotobiotic mice (51).
To date, most studies of HMO utilization by gut microbes have been performed in vitro by assessing the growth of single bacterial species in culture media containing HMO (14), or through ex vivo fermentation of HMO using fecal/intestinal microbiota from infants or animals (15, 16). Few studies investigate associations between the composition of the gut microbiota and consumed HMO profiles in vivo. A recent study by De Leoz and colleagues (18) investigated the relationship between fecal bacterial populations and HMO excreted in the stool of two BF infants. Samples were collected at birth, 1, 2 and 13–14 weeks of age. Their final time point is consistent with the 3-month sample in the current study. In both infants, they observed a shift in the fecal bacterial population from non-HMO utilizers, such as Enterobacteriaceae and Staphylococcaeae to the HMO-consumers, Bacteroidaceae and Bifidobacteriaceae (18). Relationships between fecal HMO isomers and the relative abundances of order-level bacterial taxa were determined by Pearson product-moment correlation coefficients. Consistent with our findings, both positive and negative correlations were detected between specific HMO and bacterial taxa. As an example, the relative abundance of Lactobacillales was positively correlated with the abundance of fecal MFLNH I, LnNH, pLNH and an HMO with a mass of 5130a and not negatively correlated with any of the HMO reported. Since these HMO were excreted and not utilized, this indicates that none of these HMO stimulate the growth of Lactobacillus The relative abundance of Bifidobacteriales was negatively correlated with the abundance of MFLNH I, IFLNH I, LNT, LnNH and an HMO with a mass of 5230b in feces and not positively correlated with any of the HMO reported (18). Thus, MFLNH I and LnNH had opposite effects on the relative abundance of Lactobacillales and Bifidobacteriales.
Herein, PLS regression was applied to investigate the relationship between milk HMO profiles and infant gut bacterial genus and influential HMO were identified by VIP scores. PLS regression is a supervised method that allows for the modeling of complex biological events by considering different factors at the same time (54) and is not affected by data collinearity. PLS regression coupled with VIP scores has been shown to be an excellent tool in identifying influencing variables (32, 55). Our results indicate that relative abundances of some bacterial genera detected in infant feces can be predicted by the HMO consumed. For example, the relative abundance of Bifidobacterium spp. and Bacteroides spp. in infant stool were correlated with the HMO of their mothers’ milk, which is consistent with previous culture studies showing that some strains of Bifidobacterium and Bacteroides are able to utilize HMO with high efficiency (14). In agreement with previous in vitro studies (14), the current results suggest that the relative abundances of Esherichia/Shigella, Streptococcus and Staphylococcus in infants’ feces are not associated with the HMO consumed. However, differences from previous reports were also observed in the present study. For example, in vitro culture studies have shown Enterococcus and Veillonella strains grew little or not at all when HMO was used as the carbon source (14). Results of this study demonstrate that the proportion of Enterococcus and Veillonella in infant fecal samples were associated with HMO profiles of mothers’ milk.
Those seemingly contradictory observations may be in part due to metabolic cross-feeding between members of gut bacteria. Cross-feeding is the phenomenon that metabolic products produced from one bacterial species provide substrates to support the growth of other species. Cross-feeding can result in metabolic consequences that would not be predicted simply from the substrate utilization of isolated bacteria (56). Cross-feeding has been found between strains of B. adolescentis and butyrate-producing bacteria isolated from the human gut (56). Thus, Enterococcus and Veillonella may not themselves degrade HMO, but they may be able to utilize partial breakdown products or the fermentation end products produced by other gut bacteria, such as Bifidobacterium and Bacteroides.
In conclusion, we have compared the composition of fecal microbiota between BF and FF infants by pyrosequencing of bacterial 16S rRNA genes and correlated the microbiota of BF infants with the HMO profiles consumed. Our results indicate that the fecal microbial composition of BF infants differ from that of FF infants with a higher proportion of Bacteroidetes and lower abundance of Firmicutes. Moreover, we also demonstrate that the microbial composition of BF infants is correlated with the presence of HMO in their mother’s milk. The ability of specific HMO to predict bacterial genera colonizing the infant gut should be validated in a larger cohort. If replicated, the findings would support investigating whether supplementation of infant formula with defined HMO would provide a means to selective enrich specific bacterial genera in the infant gut. (57).
Supplementary Material
What is known about this subject?
Human milk oligosaccharides (HMO) are thought to serve as the primary substrate for growth beneficial bacteria.
HMO consumption by gut bacteria has been studied in vitro.
Little is known about the association between composition of the gut microbiota and HMO profiles in vivo.
What are the new findings and/or what is the impact on clinical practice? (3–4 bullet points)
Microbial composition of breast-fed infants is positively and negatively correlated with the presence of a variety HMO in their mother’s milk and
Microbial composition of breast-fed infants can be predicted by HMO consumed.
If replicated in a larger population, the findings would support supplementation of infant formula with defined HMO as a means to enrich specific bacterial genera in the infant gut.
Acknowledgments
This study was supported by grants from National Institute of Health (R01 HD061929 and 1P30ES023512-01), a Freedom to Discover Award from the Bristol-Myers Squibb Foundation to the Division of Nutritional Sciences at the University of Illinois, Urbana, and Hatch funds (Project ILLU-971-346) distributed through the Division of Nutritional Sciences Vision 20/20 program.
We are grateful to all study subjects for participating in the study. We thank the W. M. Keck Center for Comparative and Functional Genomics at the University of Illinois for performing pyrosequencing. We also thank Mead Johnson Nutrition (Evansville, IN) for providing infant formula.
Footnotes
The authors report no conflicts of interest.
References
- 1.Rautava S, Luoto R, Salminen S, et al. Microbial contact during pregnancy, intestinal colonization and human disease. Nat Rev Gastroenterol Hepatol. 2012;9:565–76. doi: 10.1038/nrgastro.2012.144. [DOI] [PubMed] [Google Scholar]
- 2.Kull I, Melen E, Alm J, et al. Breast-feeding in relation to asthma, lung function, and sensitization in young schoolchildren. J Allergy Clin Immunol. 2010;125:1013–9. doi: 10.1016/j.jaci.2010.01.051. [DOI] [PubMed] [Google Scholar]
- 3.Li M, Wang M, Donovan SM. Early development of the gut microbiome and immune-mediated childhood disorders. Sem Reprod Med. 2014;32:74–86. doi: 10.1055/s-0033-1361825. [DOI] [PubMed] [Google Scholar]
- 4.Dominguez-Bello MG, Blaser MJ, Ley RE, et al. Development of the human gastrointestinal microbiota and insights from high-throughput sequencing. Gastroenterology. 2011;140:1713–9. doi: 10.1053/j.gastro.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Collado MC, Cernada M, Baüerl C, et al. Microbial ecology and host-microbiota interactions during early life stages. Gut Microbes. 2012;3:352–65. doi: 10.4161/gmic.21215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. [accessed 04 November 2014];Breastfeeding Report Card–United States. 2014 Internet: http://www.cdc.gov/breastfeeding/data/reportcard.htm.
- 7.Yatsunenko T, Rey FE, Manary MJ, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–7. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Palmer C, Bik EM, DiGiulio DB, et al. Development of the human infant intestinal microbiota. PLoS Biol. 2007;5:e177. doi: 10.1371/journal.pbio.0050177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartz S, Friedberg I, Ivanov I, et al. A metagenomic study of diet-dependent interaction between gut microbiota and host in infants reveals differences in developmental and immune responses. Genome Biol. 2012;13:R32. doi: 10.1186/gb-2012-13-4-r32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kunz C, Rudloff S, Baier W, et al. Oligosaccharides in human milk: structural, functional, and metabolic aspects. Annu Rev Nutr. 2000;20:699–722. doi: 10.1146/annurev.nutr.20.1.699. [DOI] [PubMed] [Google Scholar]
- 11.Ninonuevo MR, Park Y, Yin H, et al. A strategy for annotating the human milk glycome. J Agric Food Chem. 2006;54:7471–80. doi: 10.1021/jf0615810. [DOI] [PubMed] [Google Scholar]
- 12.Gnoth MJ, Kunz C, Kinne-Saffran E, et al. Human milk oligosaccharides are minimally digested in vitro. J Nutr. 2000;130:3014–20. doi: 10.1093/jn/130.12.3014. [DOI] [PubMed] [Google Scholar]
- 13.Ward RE, Niñonuevo M, Mills DA, et al. In vitro fermentation of breast milk oligosaccharides by Bifidobacterium infantis and Lactobacillus gasseri. Appl Environ Microbiol. 2006;72:4497–9. doi: 10.1128/AEM.02515-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Marcobal A, Barboza M, Froehlich JW, et al. Consumption of human milk oligosaccharides by gut-related microbes. J Agric Food Chem. 2010;58:5334–40. doi: 10.1021/jf9044205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li M, Bauer LL, Chen X, et al. Microbial composition and in vitro fermentation patterns of human milk oligosaccharides and prebiotics differ between formula-fed and sow-reared piglets. J Nutr. 2012;142:681–9. doi: 10.3945/jn.111.154427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vester Boler BM, Rossoni Serao MC, Faber TA, et al. In vitro fermentation characteristics of select nondigestible oligosaccharides by infant fecal inocula. J Agric Food Chem. 2013;61:2109–19. doi: 10.1021/jf305056f. [DOI] [PubMed] [Google Scholar]
- 17.Albrecht S, Schols HA, van den Heuvel EGHM, Voragen AGJ, Gruppen H. Occurrence of oligosaccharides n feces of breast-fed babies in their first six months of life and the corresponding breast milk. Carb Res. 2011;346:2540–50. doi: 10.1016/j.carres.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 18.De Leoz ML, Kalanetra KM, Bokulich NA, Strum JS, Underwood MA, German JB, Mills DA, Lebrilla CB. Human milk glycomics and gut microbial genomics in infant feces show a correlation between human milk oligosaccharides and gut microbiota: A proof-of-concept study. J Proteome Res. 2014 Oct 28; doi: 10.1021/pr500759e. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chapkin RS, Zhao C, Ivanov I, et al. Stool-based detection of infant gastrointestinal development using gene expression profiles from exfoliated epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2010;298:G582–9. doi: 10.1152/ajpgi.00004.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wu S, Tao N, German JB, et al. Development of an annotated library of neutral human milk oligosaccharides. J Proteome Res. 2010;9:4138–51. doi: 10.1021/pr100362f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wu S, Grimm R, German JB, et al. Annotation and structural analysis of sialylated human milk oligosaccharides. J Proteome Res. 2011;10:856–68. doi: 10.1021/pr101006u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Totten SM, Zivkovic AM, Wu S, et al. Comprehensive profiles of human milk oligosaccharides yield highly sensitive and specific markers for determining secretor status in lactating mothers. J Proteome Res. 2012;11:6124–33. doi: 10.1021/pr300769g. [DOI] [PubMed] [Google Scholar]
- 23.Yu Z, Morrison M. Improved extraction of PCR-quality community DNA from digesta and fecal samples. Biotechniques. 2004;36:808–12. doi: 10.2144/04365ST04. [DOI] [PubMed] [Google Scholar]
- 24.van den Bogert B, de Vos WM, Zoetendal EG, et al. Microarray analysis and barcoded pyrosequencing provide consistent microbial profiles depending on the source of human intestinal samples. Appl Environ Microbiol. 2011;77:2071–80. doi: 10.1128/AEM.02477-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yildirim S, Yeoman CJ, Sipos M, et al. Characterization of the fecal microbiome from non-human wild primates reveals species specific microbial communities. PLoS One. 2010;12:e13963. doi: 10.1371/journal.pone.0013963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7:335–6. doi: 10.1038/nmeth.f.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72:5069–72. doi: 10.1128/AEM.03006-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lozupone C, Knight R. UniFrac: A new phylogenetic method for comparing microbial communities. Appl Environ Microbiol. 2005;71:8228–35. doi: 10.1128/AEM.71.12.8228-8235.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang Q, Garrity GM, Tiedje JM, et al. Naive bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol. 2007;73:5261–7. doi: 10.1128/AEM.00062-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oksanen J, Blanchet FG, Kindt R, et al. Vegan: Community Ecology Package. [accessed 16 December, 2013];R package version 2.0-10. 2013 Internet: http://CRAN.R-project.org/package=vegan.
- 32.Chong IG, Jun CH. Performance of some variable selection methods when multicollinearity is present. Chemom Intell Lab Syst. 2005;78:103–12. [Google Scholar]
- 33.Vaughan EE, Schut F, Heilig HG, et al. A molecular view of the intestinal ecosystem. Curr Issues Intest Microbiol. 2000;1:1–12. [PubMed] [Google Scholar]
- 34.Yoshioka H, Iseki K, Fujita K. Development and differences of intestinal flora in the neonatal period in breast-fed and bottle-fed infants. Pediatrics. 1983;72:317–21. [PubMed] [Google Scholar]
- 35.Sela DA. Bifidobacterial utilization of human milk oligosaccharides. Int J Food Microbiol. 2011;149:58–64. doi: 10.1016/j.ijfoodmicro.2011.01.025. [DOI] [PubMed] [Google Scholar]
- 36.Adlerberth I, Wold AE. Establishment of the gut microbiota in Western infants. Acta Paediatr. 2009;98:229–38. doi: 10.1111/j.1651-2227.2008.01060.x. [DOI] [PubMed] [Google Scholar]
- 37.Veereman-Wauters G, Staelens S, Van de Broek H, et al. Physiological and bifidogenic effects of prebiotic supplements in infant formulae. J Pediatr Gastroenterol Nutr. 2011;52:764–72. doi: 10.1097/MPG.0b013e3182139f39. [DOI] [PubMed] [Google Scholar]
- 38.Hascoët JM, Hubert C, Rochat F, et al. Effect of formula composition on the development of infant gut microbiota. J Pediatr Gastroenterol Nutr. 2011;52:756–62. doi: 10.1097/MPG.0b013e3182105850. [DOI] [PubMed] [Google Scholar]
- 39.Haarman M, Knol J. Quantitative real-time PCR assays to identify and quantify fecal Bifidobacterium species in infants receiving a prebiotic infant formula. Appl Environ Microbiol. 2005;71:2318–24. doi: 10.1128/AEM.71.5.2318-2324.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jost T, Lacroix C, Braegger CP, et al. New insights in gut microbiota establishment in healthy breast fed neonates. PLoS One. 2012;7:e44595. doi: 10.1371/journal.pone.0044595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vaishampayan PA, Kuehl JV, Froula JL, et al. Comparative metagenomics and population dynamics of the gut microbiota in mother and infant. Genome Biol Evol. 2010;2:53–66. doi: 10.1093/gbe/evp057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mazmanian SK, Liu CH, Tzianabos AO, et al. An immunomodulatory molecule of symbiotic bacteria directs maturation of the host immune system. Cell. 2005;122:107–18. doi: 10.1016/j.cell.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 43.Comstock LE. Importance of glycans to the host-bacteroides mutualism in the mammalian intestine. Cell Host Microbe. 2009;5:522–6. doi: 10.1016/j.chom.2009.05.010. [DOI] [PubMed] [Google Scholar]
- 44.Marcobal A, Sonnenburg JL. Human milk oligosaccharide consumption by intestinal microbiota. Clin Microbiol Infect. 2012;18 (Suppl 4):12–5. doi: 10.1111/j.1469-0691.2012.03863.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hamer HM, Jonkers D, Venema K, et al. Review article: the role of butyrate on colonic function. Aliment Pharmacol Ther. 2008;27:104–19. doi: 10.1111/j.1365-2036.2007.03562.x. [DOI] [PubMed] [Google Scholar]
- 46.Ley RE, Turnbaugh PJ, Klein S, et al. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–3. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 47.Nadal I, Santacruz A, Marcos A, et al. Shifts in clostridia, bacteroides and immunoglobulin-coating fecal bacteria associated with weight loss in obese adolescents. Int J Obes (Lond) 2009;33:758–67. doi: 10.1038/ijo.2008.260. [DOI] [PubMed] [Google Scholar]
- 48.Grummer-Strawn LM, Mei Z Centers for Disease Control and Prevention Pediatric Nutrition Surveillance System. Does breastfeeding protect against pediatric overweight? Analysis of longitudinal data from the Centers for Disease Control and Prevention Pediatric Nutrition Surveillance System. Pediatrics. 2004;113:e81–6. doi: 10.1542/peds.113.2.e81. [DOI] [PubMed] [Google Scholar]
- 49.Martín-Sosa S, Martín MJ, García-Pardo LA, et al. Sialyloligosaccharides in human and bovine milk and in infant formulas: variations with the progression of lactation. J Dairy Sci. 2003;86:52–9. doi: 10.3168/jds.S0022-0302(03)73583-8. [DOI] [PubMed] [Google Scholar]
- 50.Asakuma S, Hatakeyama E, Urashima T, et al. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J Biol Chem. 2011;286:34583–92. doi: 10.1074/jbc.M111.248138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marcobal A, Barboza M, Sonnenburg ED, et al. Bacteroides in the infant gut consume milk oligosaccharides via mucus-utilization pathways. Cell Host Microbe. 2011;10:507–14. doi: 10.1016/j.chom.2011.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sela DA, Chapman J, Adeuya A, et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc Natl Acad Sci U S A. 2008;105:18964–9. doi: 10.1073/pnas.0809584105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.LoCascio RG, Ninonuevo MR, Freeman SL, et al. Glycoprofiling of bifidobacterial consumption of human milk oligosaccharides demonstrates strain specific, preferential consumption of small chain glycans secreted in early human lactation. J Agric Food Chem. 2007;55:8914–9. doi: 10.1021/jf0710480. [DOI] [PubMed] [Google Scholar]
- 54.Palermo G, Piraino P, Zucht HD. Performance of PLS regression coefficients in selecting variables for each response of a multivariate PLS for omics-type data. Adv Appl Bioinforma Chem. 2009;2:57–70. doi: 10.2147/aabc.s3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mehmood T, Martens H, Sæbø S, et al. A partial least squares based algorithm for parsimonious variable selection. Algorithms Mol Biol. 2011;6:27. doi: 10.1186/1748-7188-6-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Belenguer A, Duncan SH, Calder AG, et al. Two routes of metabolic cross-feeding between Bifidobacterium adolescentis and butyrate-producing anaerobes from the human gut. Appl Environ Microbiol. 2006;72:3593–9. doi: 10.1128/AEM.72.5.3593-3599.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chichlowski M, German JB, Lebrilla CB, et al. The influence of milk oligosaccharides on microbiota of infants: Opportunities for formulas. Annu Rev Food Sci Technol. 2011;2:331–51. doi: 10.1146/annurev-food-022510-133743. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.