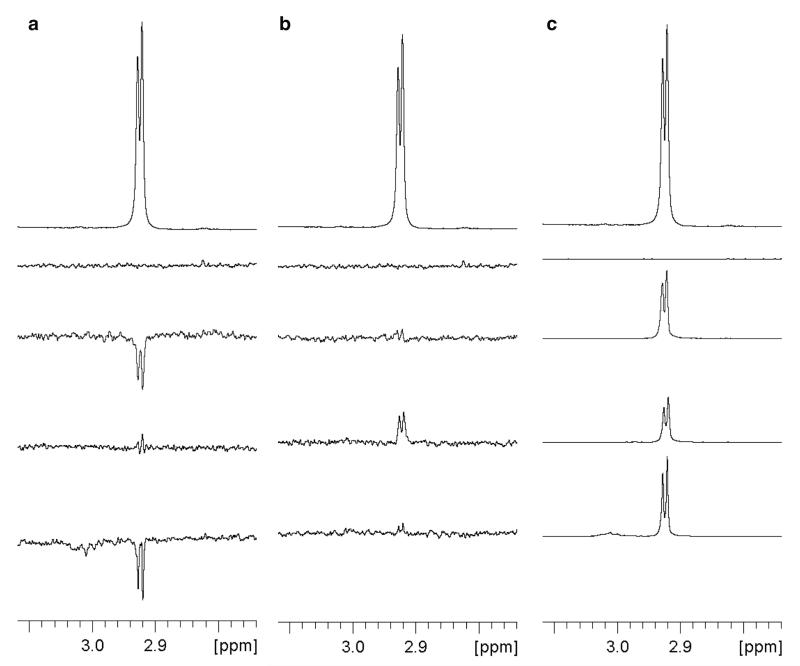
Fig. 8.
Identification of ligand binding to a protein using 1D 1H ligand-observed NMR spectroscopy. (a) WaterLOGSY, (b) STD, and (c) relaxation-edited binding and displacement experiments are shown. From top to bottom: normal 1H spectrum of ligand in the absence of protein; a control spectrum of buffer alone; ligand in the absence of protein; ligand in the presence of protein; ligand in the presence of protein and a known high affinity binder (a “displacer”). The spectra show a doublet from a methyl group adjacent to an amide NH of a fragment ligand binding to the human bromodomain of BAZ2B protein (MW = 13.6 kDa, which is on the lower limit for ligand-based NMR techniques), displaced by a high-affinity peptide H3Kac14 (49). The restoration of a spectrum similar to ligand alone by addition of the displacer demonstrates that the ligand binds at the same site. A subsequent X-ray crystal structure of the protein-fragment complex demonstrated that the fragment bound at the Kac binding site of the bromodomain