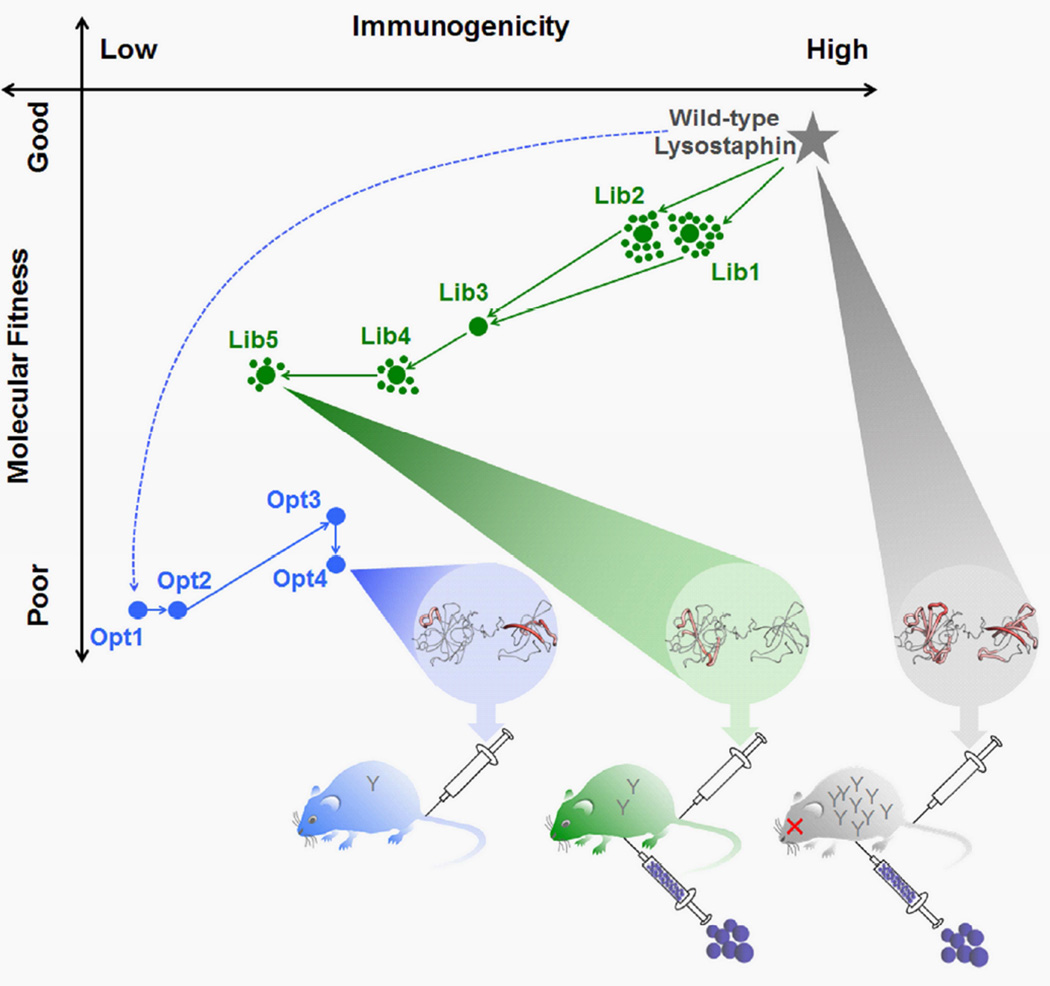
Fig. 1. Schematic overview of molecular engineering process and in vivo outcomes.
(Top) A plot of Molecular Fitness vs. Immunogenicity is shown for individually optimized variants and selected library members. The Y-axis reflects trends in experimental performance measurements and the X-axis reflects trends in computational predictions of T cell epitope content. LSTWT has both high fitness and high immunogenicity, where the latter is shown as a structural epitope map (grey) (thick red indicates dense epitope content and thin grey indicates no epitope content). Deimmunization by design of individual optimal variants is shown in blue, where the initial design was fully depleted of DR4 epitopes, but failed to express. Subsequent designs reverted problematic mutations in a reverse engineering process, ultimately yielding the expressible, aglycosylated, but inactive variant Opt4, whose structural epitope map is shown (blue). Deimmunization by library design and screening is shown in green. Deimmunizing mutations were accumulated in an iterative process, ultimately yielding the active and deimmunized Lib5 variant, whose structural epitope map is shown (green). (Bottom) The in vivo immunogenicities of LSTWT, Opt4, and Lib5 were evaluated in transgenic DR4 mice, and trends in observed antibody titers are indicated as Y-shaped IgG cartoons in the mice. Subsequently, the efficacy of LSTWT and Lib5 were evaluated in a recurrent bacteremia model that required repeated dosing of the antibacterial enzyme. Variant Lib5 enabled repeated rescue of DR4 mice, whereas LSTWT lost efficacy due to high antibody titers. See also Table S1.