Figure 2.
DEER distances of inactive and fully active β2AR. See Figure S2 for raw DEER data.
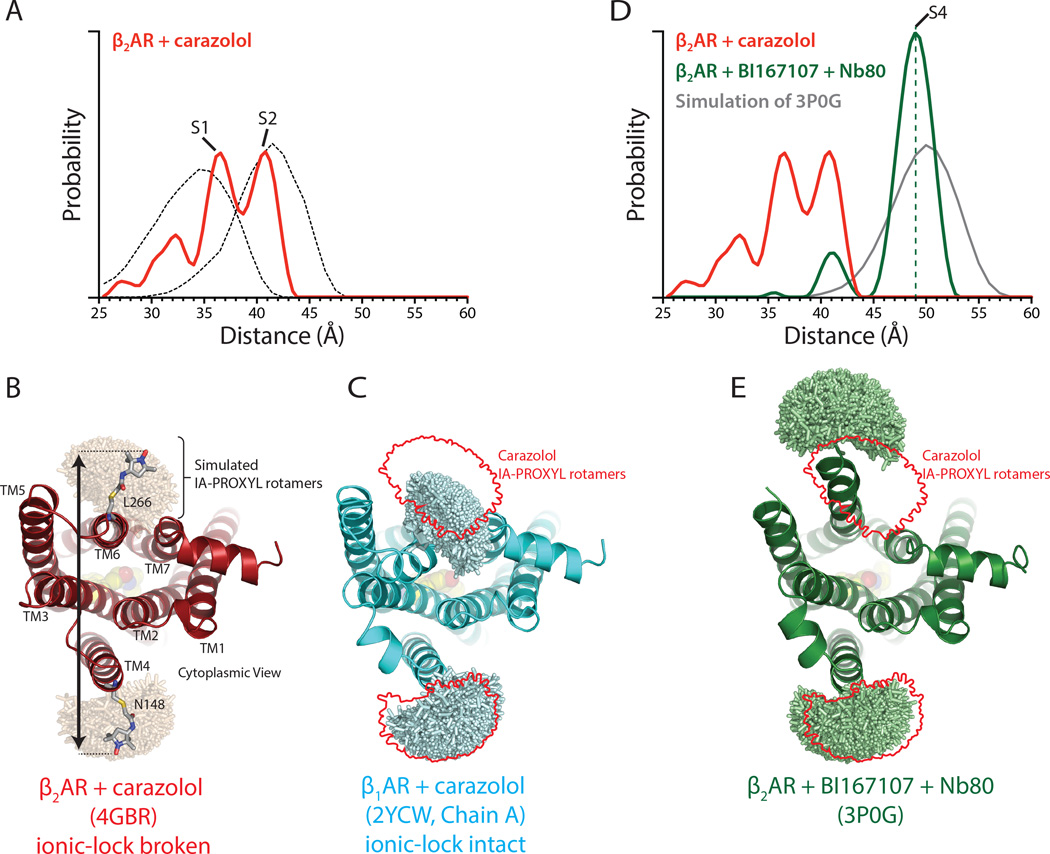
(A) Distance distribution for carazolol-bound β2AR. The dotted lines show simulated nitroxide spin label distance distributions for a state with a broken ionic lock using carazolol-bound β2AR (PDB ID: 4GBR), and a state with an ionic lock intact using β1AR (PDB ID: 2YCW) as shown in panels B and C.
(B) IA-PROXYL rotamers modeled onto β2AR bound to carazolol (PDB ID: 4GBR) using MTSSLWizard. The distance between these possible rotamers is then determined in a pairwise manner to yield the predicted distance distribution shown in A.
(C) Similar analysis as in B was performed on the structure of β1AR bound to carazolol but with an intact ionic lock. The red outline indicates rotamers modeled in B. The mean distance for a state with an intact ionic lock is predicted to be shorter than for β2AR with a broken ionic lock.
(D) Distance distribution for β2AR bound to BI167107 and Nb80. The dashed green line serves as a marker for the S4 state. The grey line represents the simulated distance distribution for β2AR bound to BI167107 and Nb80 using the previously determined crystal structure (PDB ID: 3P0G) as shown in panel E.
(E) IA-PROXYL rotamer modeling of activated β2AR bound to BI167107 and Nb80.