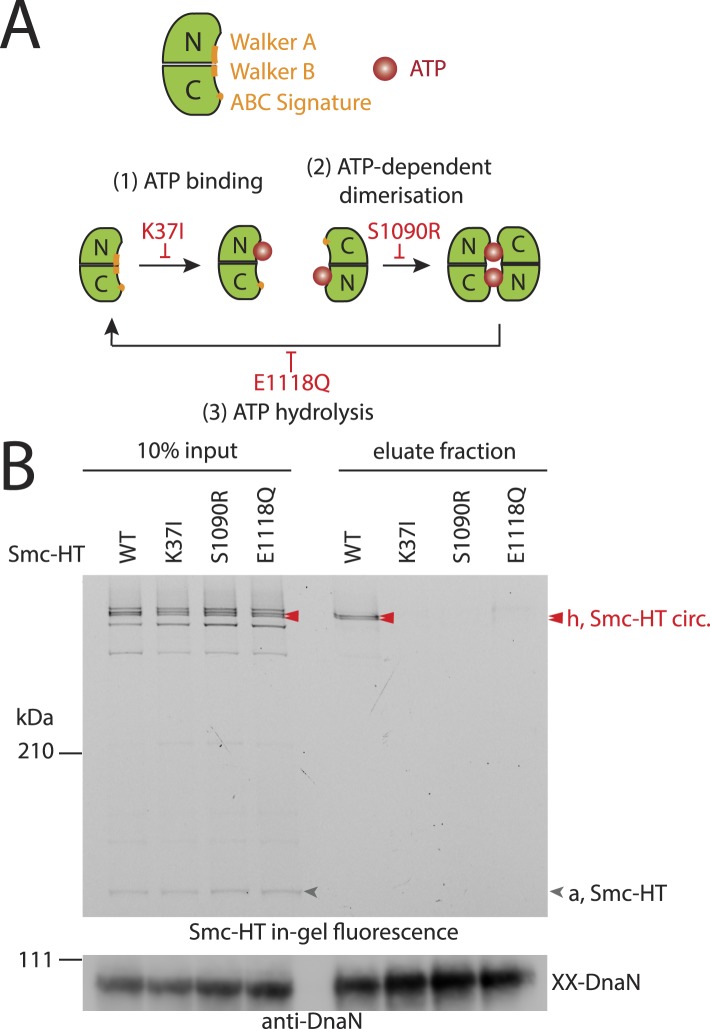
Figure 3. The Smc ATPase is required for loading of DNA into Smc–ScpAB.
(A) A scheme for the ATP hydrolysis cycle of Smc. Schematic positions for Walker A, Walker B and ABC-signature motifs on the Smc head domain are shown (top row). ATP binding to the Walker A domain is blocked in Smc(K37I) ‘(1)’. ATP-dependent engagement of two Smc heads is abolished in the Smc(S1090R) mutant ‘(2)’. The E1118Q mutation strongly reduces ATP hydrolysis ‘(3)’. (B) Smc ATPase mutations abolish chromosomal loading of Smc–ScpAB. In-gel fluorescence detection of Smc-HT of input and eluate fractions from a representative chromosome entrapment assay performed with strains BSG1782 and BSG1784-6. Protein extracts (10% of input) were loaded next to samples subjected to the entrapment assay. Selected cross-linked species of Smc-HT are labeled (top panel). Detection of cross-linked species of DnaN by immunoblotting was used as internal assay control (bottom panel). The following figure supplement is available: Figure 3—figure supplement 1: ATPase mutants of Smc–ScpAB.