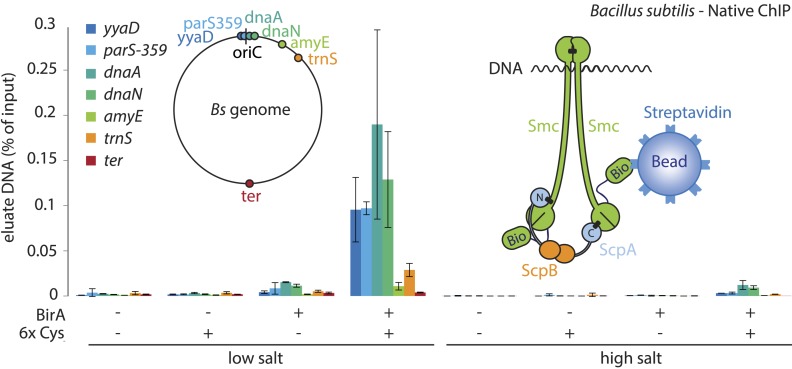
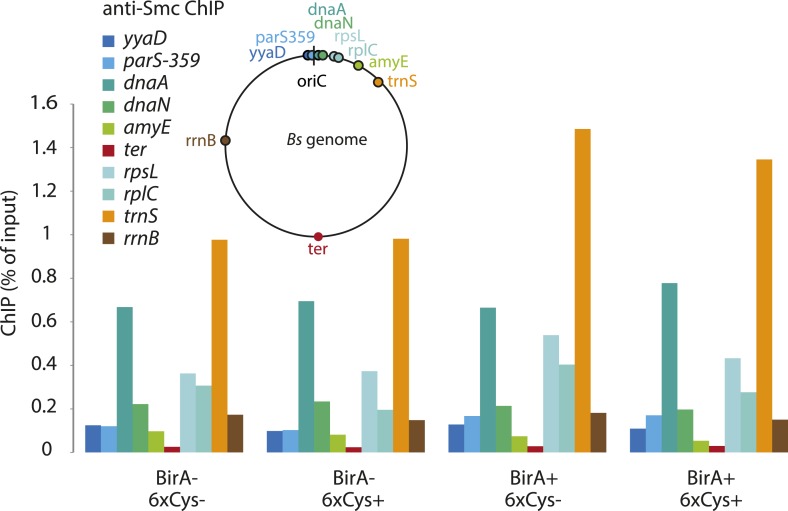
Figure 5. A physical interaction of Smc–ScpAB rings and chromosomal DNA.
Co-purification of chromosomal DNA fragments with native Smc–ScpAB. Cells of strains BSG1104-5 and BSG1107-8 were treated with the cross-linker BMOE prior to cell lysis. Strains carrying (‘+’) or lacking (‘−’) cysteine mutations (‘6xCys’) in the Smc-AviTag construct were expressed in presence (‘+’) or absence (‘−’) of the biotin ligase (‘BirA’). Beads were washed in the presence of either a 150 mM ammonium acetate buffer (‘low salt’) or a 2 M sodium chloride buffer (‘high salt’). The co-purification of DNA fragments with Smc-biotin on streptavidin beads was measured by quantitative PCR using primer pairs specific for genomic positions indicated on a representation of the circular B. subtilis genome. Mean values and standard deviations were calculated from two independent biological replicates. The following figure supplement is available: Figure 5—figure supplement 1: Chromatin immuno-precipitation of Smc.