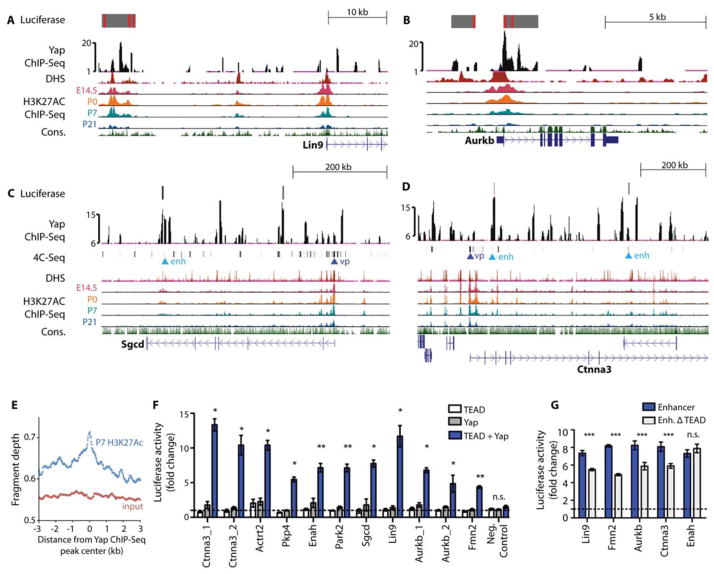
Figure 2. Preferential expression of Yap target genes in the fetal heart.
(A–D) Genome browser view of Yap ChIP-Seq enriched peaks for the labeled genes. Alignments are shown for stage-specific H3K27Ac ChIP-Seq, DHS, and 4C anchor points (for Ctnna3 and Sgcd). For 4C, “vp” denotes viewpoint and “enh” denotes enhancer. Grey blocks show regulatory regions used as luciferase reporters with red lines indicating consensus Tead binding motif. The y-axes of Yap ChIP peaks show the normalized read number. (E) Quantification of P7 heart H3K27Ac ChIP-Seq reads in the 6-kb range around Yap ChIP-Seq peaks. N=2 biologic replicates. (F) Luciferase reporter assay data showing that Yap/Tead co-activated target gene expression through regulatory regions identified in Yap ChIP-Seq. N=3 independent transfection experiments. *P<0.05; **P<0.01; ***P<0.001. (G) Luciferase reporter assay data of Yap enhancers with or without TEAD motifs. N=3 independent transfection experiments. All luciferase constructs were co-transfected with Yap and TEAD expression vectors. ***p<0.001 (Mann-Whitney), n.s. not significant. Error bars are standard deviations. Activity was normalized to that of cells expressing the pGL3 vector.