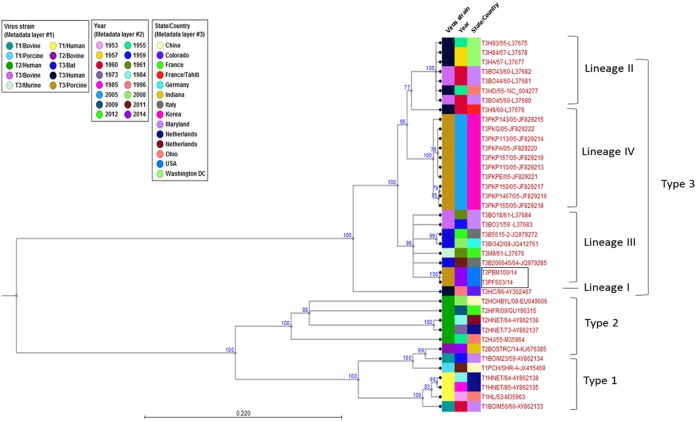
FIG 4 .
Phylogenetic analysis of novel orthoreoviruses. Shown is evolutionary analysis for tree topologies of the complete MRV3 S1 segment open reading frames constructed through the maximum likelihood method using the Jukes-Cantor evolutionary model in CLC workbench 7.0.4. Branches with less than 75% bootstrap support were collapsed. All MRV3 strains are grouped into lineages I to IV. Branches of FS03 and BM100 within lineage III are boxed. The metadata layers, including virus strain, isolation year, and place of origin, are color coded. The scale bar shows the evolutionary distance of 0.220 substitution per site. The tree calculations were unrooted.