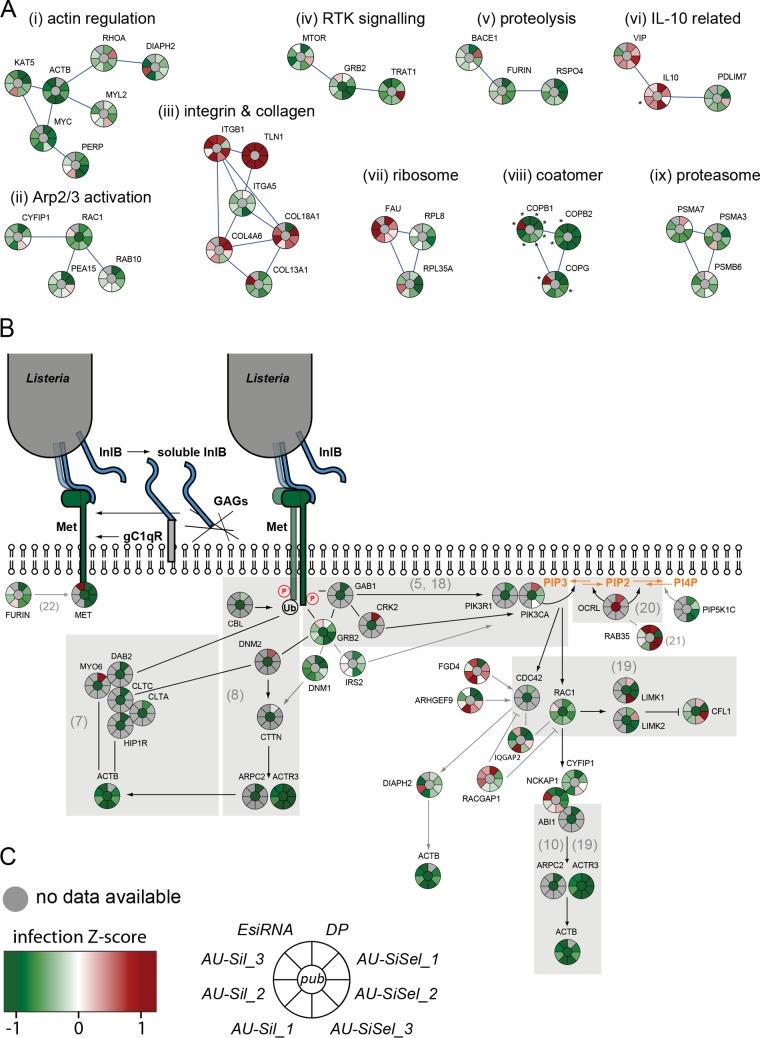
FIG 3 .
Results from validation screens for Listeria infection. (A) Gene clusters (STRING stringency, 0.7) of functional groups obtained with all hits that gave consistent results for infection with no more than one outlier out of 7 or 8 individual siRNA sequences or pools: (i) action-related cluster, (ii) RAC1 and Arp2/3 activation-related cluster, (iii) integrin/collagen-related cluster, (iv) receptor tyrosine kinase (RTK) signaling and GRB2-related cluster, (v) proteolytic cleavage, (vi) genes connected to interleukin-10 (IL-10), (vii) ribosome-related cluster, (viii) coatomer-related cluster, and (ix) proteasome-related cluster. Knockdowns that resulted in cell numbers lower than 500 are marked with asterisks. (B) Validation results for genes that have been shown to be involved in Met/InlB-dependent Listeria invasion (gray background) or that can be associated with the Met/InlB pathway (STRING stringencies of 0.235 for furin/MET and 0.4 for other connections). References are indicated in parentheses. (C) Infection Z score color code and diagram depicting the expectation according to published results (center) as well as results obtained with the Dharmacon pooled genome-wide libraries (DP), with the Ambion silencer select unpooled siRNAs (AU-SiSel_1 to -3), the Ambion silencer unpooled siRNAs (AU-Sil_1 to -3), and the Sigma esiRNA pooled siRNAS (EsiRNA).