FIG 7.
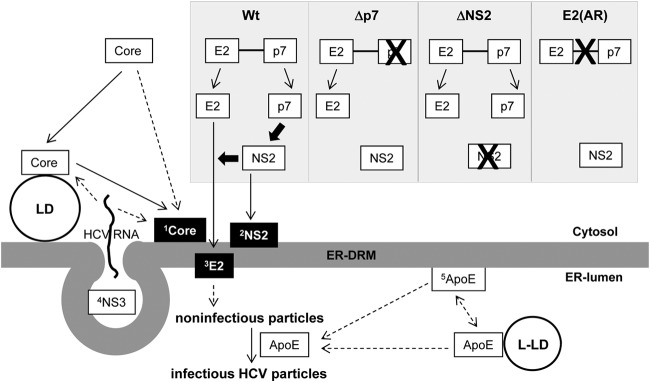
Model for the role of DRM targeting of viral and host proteins during HCV particle assembly. 1Core associates with the DRM (Fig. 1) (30, 34). Core could be targeted to the DRM directly or indirectly following its initial interaction with LD (11, 20, 53, 68). 2NS2 was targeted to the DRM in a p7 (and therefore E2-p7 processing)-dependent manner (Fig. 3A, 4A, and B). 3E2 was targeted to the DRM in an NS2-dependent manner (Fig. 4C). Defects in p7, NS2, or E2-p7 processing block both NS2 and E2 localization to the DRM and virus assembly (Fig. 1, 3, and 4). 4NS3 and other nonstructural proteins (NS4A, NS4B, NS5A, and NS5B) comprising the replication complexes (RC) were targeted to the DRM for HCV RNA replication (27, 28). RC are likely enclosed by membrane invaginations called membranous web (69–71). Positive-strand HCV RNA produced by RC will then interact with Core for encapsidation. Currently, it is unclear whether HCV RNA interacts with Core on the LD, at the DRM or both (50). Encapsidated Core will then interact with envelope proteins (72), and this interaction will likely trigger enveloped particle budding at the ER-DRM into the ER-lumen. 5ApoE incorporation to the enveloped particles (31, 58), potentially through its interaction with envelope proteins (60, 63), is critical for HCV infectivity (62, 64). We speculate that ER-DRM-enriched ApoE facilitates this process. However, further study will be needed to clarify this issue. Solid arrows indicate the pathways supported by experimental evidence from our data and previous literature. Dashed arrows indicate potential or undefined pathways. Thick black arrows indicate the driving forces for NS2 or E2 localization to the DRM. The selective MβCD sensitivity of DRM-associated Core, E2, and NS2 is indicated by a black background box. L-LD, luminal LD.
