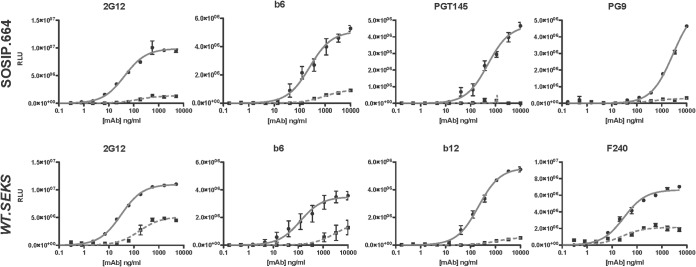
FIG 6.
The binding of MAbs to Env trimers is chaotrope sensitive. The MAbs were titrated against D7324-captured, nativelike BG505 SOSIP.664-D7324 trimers (top row) or uncleaved, nonnative BG505 WT.SEKS-D7324 gp140 proteins (bottom row). Bound MAb, detected as luminescence (RLU), is expressed on the y axis as a function of MAb concentration (ng/ml) on the x axis. The MAb binding after chaotrope (NaSCN) or control (PBS) treatment is represented by dashed and solid lines, respectively. Note the differences in y-axis scale between diagrams. The data points are the mean results from three intra-assay replicates, and the error bars show the SEM. Each plot is representative of ≥2 replicate experiments.