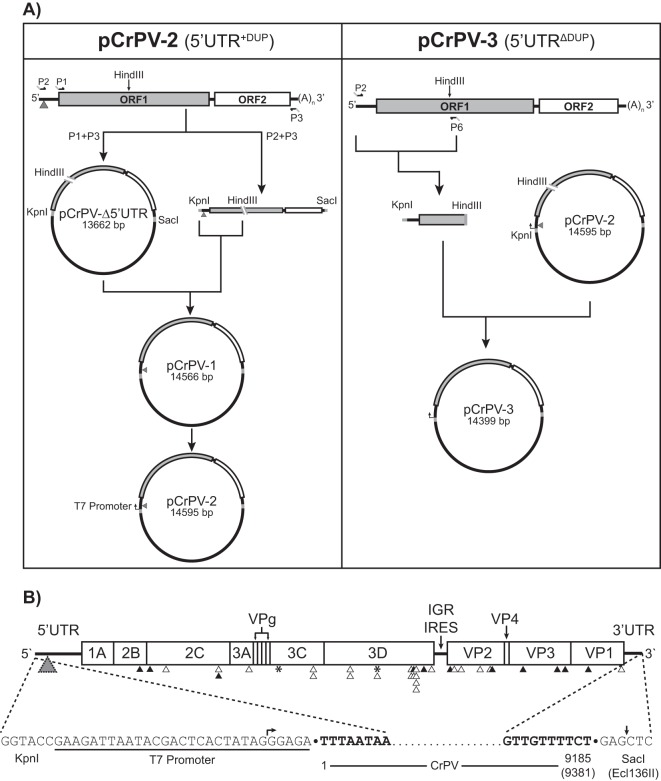
FIG 3.
Construction and map of the full-length cDNA clones CrPV-2 and CrPV-3. (A) RNA from CsCl-purified CrPV was reverse transcribed using an oligo(dT) primer. Primers P1 and P3 were used to clone the CrPV genome without the 5′ UTR into pAC5.1/V5 His B while primers P2 and P3 containing KpnI and SacI restriction sites, respectively, were used to amplify the CrPV genome. CrPV contains an endogenous HindIII site in ORF1. Therefore, to clone the 5′ UTR, both the P1/P2 amplicon and pCrPV-Δ5′ UTR were digested with HindII and KpnI. The resulting fragments were ligated together to generate pCrPV-1. A T7 polymerase promoter was added to pCrPV-1 via primer incomplete polymerase extension (PIPE) to create pCrPV-2. To create pCrPV-3, a PCR fragment derived from the 2006 stock of CrPV was inserted into the KpnI and HindIII sites of pCrPV-2 followed by a T7 promoter. (B) At top the organization of the CrPV-2 and CrPV-3 cDNA genomic sequences showing all annotated proteins is shown. Arrowheads indicate either synonymous (black) or nonsynonymous (white) changes that differ from the published CrPV sequence (GenBank accession number NC_003924). Insertion-deletion events are indicated by an asterisk. A gray triangle indicates the location of the 196-nt duplication in CrPV-2. See Table 1 for specific nucleotide changes. The sequences of the 5′ and 3′ ends of the cloned viral genomes are shown at the bottom of the panel. Nucleotides corresponding to the viral genome are bolded. Arrows indicate the beginning and end of the in vitro transcript obtained from using T7 polymerase with Ecl136II-linearized plasmid.