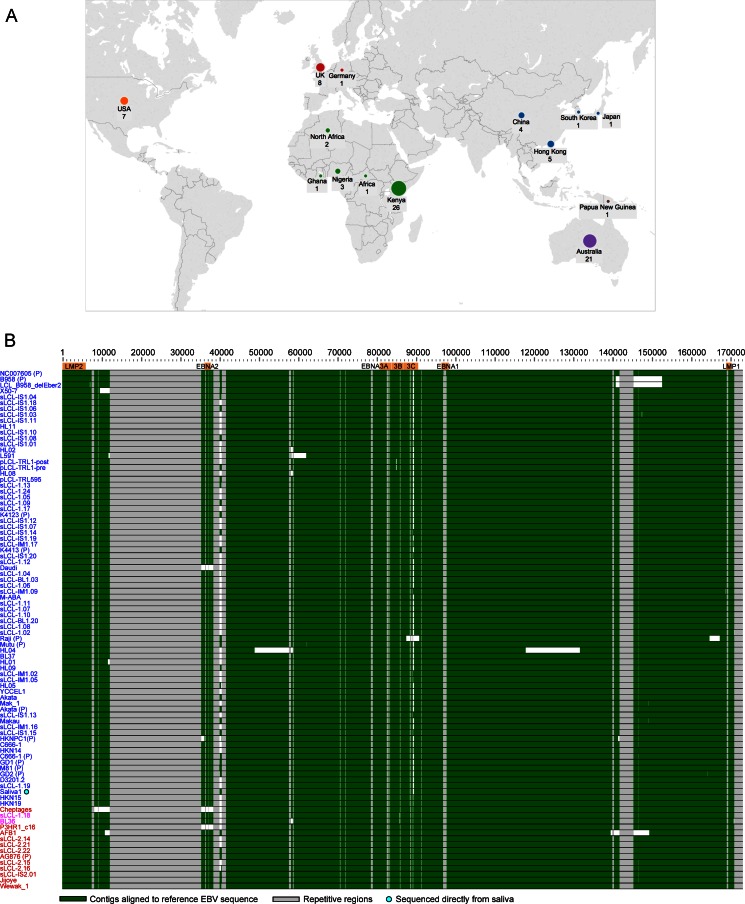
FIG 1.
Origin of samples and alignment of new EBV genome sequences. (A) World map depicting the origins of the 83 unique EBV genomes sequenced and analyzed in this study. The sizes of the circles are proportional to the number of samples from each geographic region, with country and number of genomes annotated. One sample without country information is listed as “African,” and one sample of unknown geographic origin is not shown. (B) Whole-genome multiple-sequence alignment of all EBV genomes, including the EBV (B95-8/Raji) reference genome (NC_007605, top) and 12 previously published genomes (annotated with a “P”). Strains are ordered by their similarity to the sequence of the type 1 reference strain (NC_007605), shown at the top. Type 1 genomes are in blue, type 2 genomes are in red, and recombinant (type 1/2) sequences are in purple. Contigs are indicated in green. Repetitive regions are masked out in gray. Gaps and deletions are in white. The wild-type genome derived from the saliva of a healthy individual is annotated with a turquoise dot. The HL04 Hodgkin lymphoma biopsy specimen has an inversion of a section of the genome from position 65000 to 125000. Known deletions in Daudi, P3HR1, pLCL-TRL1, and an LCL generated with a B95-8-based BAC (LCL delEBER2) were detected.