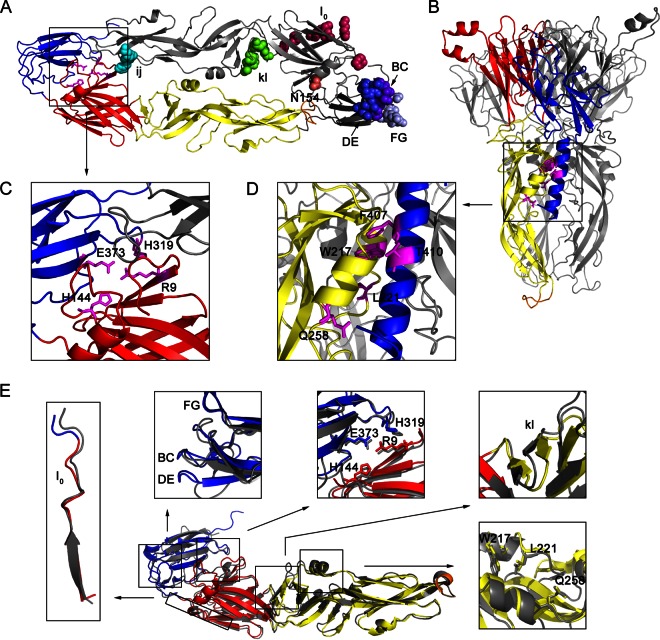
FIG 1.
Several amino acids in the E protein potentially involved in viral entry (Table 1). (A) Top view of the dimeric prefusion E protein ectodomain conformation of strain AT31, a homology model of the crystal structure of Protein Data Bank identification number (PDB ID) 3P54. In the ribbon diagram of the bottom E protein monomer, DI, DII, DIII, and the fusion peptide loop are shown in red, yellow, blue, and orange, respectively, and stick representations of four amino acids, R9, H144, H319, and E373, participating in the DI-DIII interaction are shown in magenta. In the ribbon diagram of the top E monomer, shown in gray, several amino acids in the receptor-binding motif or critical for membrane fusion are indicated by colored spheres. Residue 154 lacks a carbohydrate modification because the E ectodomain was purified from bacterial inclusion bodies in a previous study (16). (B) Side view of trimeric postfusion E protein ectodomain conformation, a homology model of the PDB ID 4FG0 docked H1 helix by Zdock server. The H1 helix is shown in blue. Only one E monomer is colored, and the others are shown in gray. Stick representations of five amino acids, W217, L221, Q258, F407, and T410, participating in the three-helix interaction of aA, aB, and H1 are shown in magenta. (C and D) Enlargement of the DI-DIII interaction and zippering reaction, respectively. (E) Structural comparison of the prefusion E conformation of JEV (PDB ID 3P54) and TBEV (PDB ID 1SVB). The three domains and fusion peptide of the E monomer of JEV are colored as described in panel A, and the E monomer of TBEV is shown in gray. The comparisons of a selected region or amino acids for mutagenesis are shown close to the E monomer. Structural comparisons of the prefusion E protein of JEV with other flaviviruses, WNV (PDB ID 2I69) and DENV (PDB ID 1OKE), are not shown.