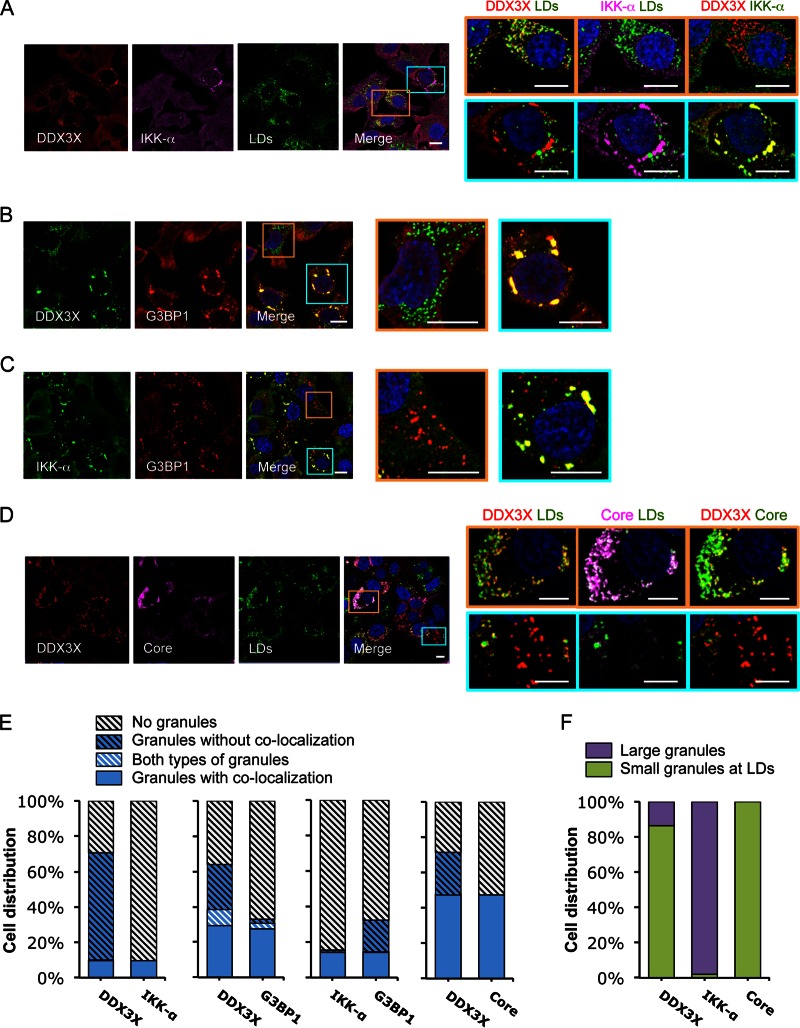
FIG 2.
HCV infection induces formation of two subsets of DDX3X granular structures with distinct subcellular localizations. Huh-7.5.1 cells were infected with JFH-1 HCVcc for 2 days. Various cellular proteins and organelles then were detected by immunofluorescence and confocal microscopy. Scale bars, 10 μm. (A to D) Magnifications of two independent selected areas are shown on the right: small DDX3X-HCV core protein granules at the LD surface (orange area) and large DDX3X–IKK-α granules (blue area). (A) DDX3X with IKK-α or LDs. (B) DDX3X with SGs. (C) IKK-α with SGs. (D) DDX3X, HCV core protein, and LDs. (A and D) Colors in images on the far right were changed for better visualization of IKK-α (A) or HCV core protein (D), shown in green instead of magenta. (E) Quantification of cell distribution of colocalizing granular structures positive for various combinations of proteins. Confocal microscopic images from each comparison group (10 to 25 randomly selected fields from three independent experiments performed for panels A to D, at least 120 cells per group) were counted. The comparison groups are DDX3X with IKK-α (far left), DDX3X with G3BP1 (second from the left), IKK-α with G3BP1 (second from the right), and DDX3X with HCV core protein (far right). Results shown are percentages of cells without any granular structures (light gray striped bars) or those with granular structures either colocalized with each other (blue bars) or not colocalized (blue striped bars). Blue-white striped bars represent percentages of cells with both colocalized and noncolocalized granules. (F) Quantification of cell distribution containing large granules (purple bars) or small granules that colocalize with LDs (green bars) stained positive for DDX3X, IKK-α, or HCV core protein. At least 40 cells for each protein were counted, and percentages of cells with each pattern of granular structure are shown.