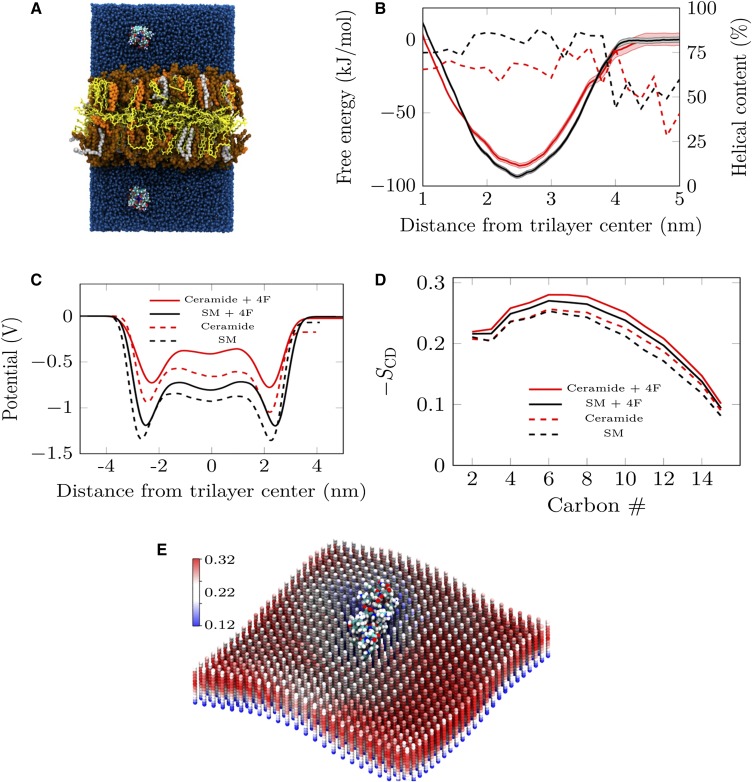
Fig. 5.
Atomistic simulations showing that 4F alters the properties of the surface monolayer in protein-free LDL. A: Side view of the initial structure of one replica of the Helix_trilayer simulations with ceramide. Two 4F peptides are positioned in the water phase in the beginning of the simulation. Cholesteryl oleate is shown in yellow, POPC in brown, cholesterol in orange, and ceramide in gray. Water is shown in blue, and the peptides are depicted with standard atom coloring (carbon, cyan; oxygen, red; hydrogen, white; and nitrogen, blue). B: The free energy profiles for the 4F peptide binding to the two trilayer surfaces. Black curves stand for the system with SM (prior to SMase treatment), while red ones show data for the system with ceramide (after SMase treatment). The shaded area shows the statistical error estimated with the bootstrapping analysis. The dashed lines show the average helical contents of the peptide in the umbrella windows. C: The electrostatic potential profile across the lipid trilayer system. Red and black curves show data for trilayers containing ceramide and SM, respectively. Dashed lines show results without the bound peptides (one in each leaflet), while solid lines stand for systems with the peptide. D: Simulation data for the deuterium order parameter −SCD, characterizing conformational order in the hydrocarbon chain region. Results are given for the saturated palmitate chains of POPC with the carbon numbering starting from the carbonyl carbon. Coloring as in C. E: The −SCD order parameter values of the palmitate chain of POPC given on a two-dimensional grid. Here, the location of the peptide was kept constant during the analysis to see the local effects induced by 4F binding. Each column represents data averaged in one grid point. The 14 spheres in each column represent the average position of the representative carbon (2nd to 15th carbon in a saturated palmitate chain of POPC), whereas their color represents the −SCD value. The lattice structure shown for lipid positions is due to the lipid order depicted here on a grid and is thus due to visualization only. In practice, the simulated surface monolayers are always in a fluid phase.