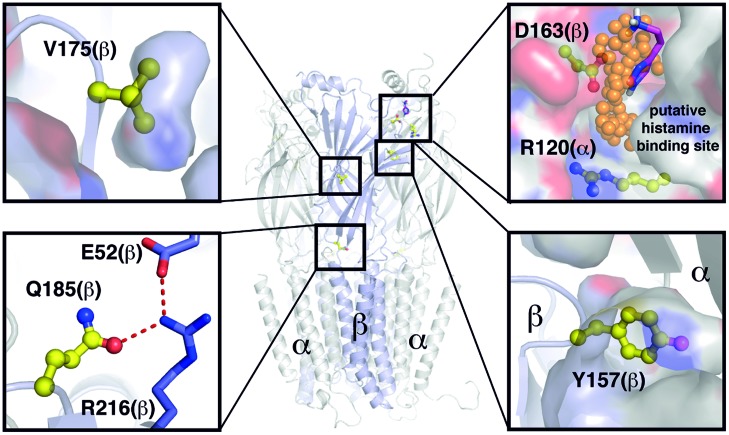
FIGURE 7.
Overview of positions and functions of mutated residues. Structural model based on PDB ID 4COF (Miller and Aricescu, 2014). Backbone displayed as cartoon, van der Waals surface as surface, selected residues as sticks, or balls and sticks, respectively. Extracellular domains visible as sheets, transmembrane domains as helical domains. Putative binding positions of small organic molecules according to TRIDOCK analysis (te Heesen et al., 2007) as orange spheres and histamine in magenta. Y157 is actively taking part in formation of the interface between the two subdomains. D163(β) and R120(α) form a putative histamine binding site at the interface between subdomains α and β. Q185 at the interface between transmembrane and extracellular domains is part of a hydrogen bonding network with the highly conserved residues E52 and R216. The V175 side chain is buried within the extracellular subdomains, and thus important for their correct folding. None of the investigated residues except D163(β) and R120(α) show a putative small molecule binding site.