Abstract
Background: MiR-126 is frequently downregulated in a variety of malignancies and acts as a potential tumor suppressor. It also played a tumor suppressor role in human melanoma through the direct or indirect repression of several key oncogenic molecules. Methods: qRT-PCR assay was performed to examine the expression of miR-126. Associations between miR-126 expression and various clinicopathological characteristics were analyzed using the χ2 test. Survival rate was determined with Kaplan-Meier and statistically analyzed with the log-rank method between groups. Survival data were evaluated through multivariate Cox regression analysis. Results: Significant differences for miR-126 expression were shown between dysplastic nevi and primary cutaneous melanoma (P<0.01), between primary melanoma and metastatic cutaneous melanomas (P<0.01), and between primary cutaneous melanomas and metastatic cutaneous melanomas (P<0.001). The patients with low miR-126 expression showed shorter 5-year overall survival than those with high miR-126 expression (P=0.039; log-rank test). Multivariate regression analysis showed that the status of miR-126 expression was an independent prognostic factor overall survival (HR=3.782, 95% CI: 2.479-16.334, P=0.005). Conclusion: The status of miR-126 might be an independent prognostic factor for patients with cutaneous melanoma.
Keywords: microRNA-126, prognosis, cutaneous melanoma, biomarker
Introduction
The incidence of cutaneous melanoma has risen considerably during recent decades around the world [1,2]. Malignant melanoma is highly characteristic of aggressive invasion, early metastasis and resistance to chemotherapy or radiotherapy, which results in the increased incidence and mortality worldwide, especially among white populations during past decades [3]. Therefore, better understanding of the molecular mechanisms about malignant melanoma tumorigenesis and progression will be helpful to explore novel therapeutic agents and prognostic markers in the treatment of patients with cutaneous melanoma.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that control many fundamental biological processes [4]. miRNAs exert their important regulatory functions via negatively regulating specific target gene expression posttranscriptionally [5]. Increasing evidence revealed the relationship between cancer pathology and miRNA expression alterations.
MicroRNA-126 (miR-126), a key positive regulator, promotes angiogenesis in response to angiogenic growth factors, such as vascular endothelial growth factor-A (VEGF-A). This is done by repressing negative regulators of signal transduction pathways [6]. In this context, miR-126 works as an oncogene, but several studies have shown that miR-126 is down-regulated in different malignancies and is a potential tumor suppressor [7-10]. Previously, Felli et al demonstrated that miR-126 played a tumor suppressor role in human melanoma through the direct or indirect repression of several key oncogenic molecules [11]. However, the clinical significance of miR-126 in cutaneous melanoma has not been reported.
Materials and methods
Study population and tissue samples
A total of 16 cases of dysplastic nevi, 18 cases of melanoma metastases and 108 cases of primary cutaneous melanoma tissue samples were collected directly from surgery after removal of the necessary amount of tissue for routine pathology examination in the Department of Pathology, Union Hospital, Tongji Medical College, Huazhong University of Science and Technology between May 2005 and December 2013. The histological diagnosis, Breslow thickness and Clark level were re-examined from 1 to 5 original sections of the primary tumor by the same pathologist who was unaware of the clinical data. The tumors were frozen at -80°C in a guanidinium thiocyanate solution and RNA was extracted from the samples using a standard Trizol RNA extraction method (Invitrogen, Life Technologies, Carlsbad, CA, USA). The study has been approved by the local Ethical Committee. Informed written consent was obtained from all patients. The characteristics of patients are shown in Table 1.
Table 1.
Tissue miR-126 expression level and clinicopathological factors in 108 patients with primary cutaneous melanomas
| miR-126 expression level | P value | |||
|---|---|---|---|---|
|
|
||||
| Variables | Case (n) | Low (n=55) | High (n=53) | |
| Age | ||||
| ≤55 | 37 | 21 | 16 | 0.422 |
| >55 | 71 | 34 | 37 | |
| Gender | ||||
| Male | 58 | 27 | 31 | 0.342 |
| Female | 50 | 28 | 22 | |
| Thickness (mm) | ||||
| ≤2.0 | 42 | 16 | 26 | 0.048 |
| >2.0 | 66 | 39 | 27 | |
| Ulceration | ||||
| Absent | 31 | 7 | 24 | <0.001 |
| Present | 77 | 48 | 29 | |
| Histologic type | ||||
| SSM | 55 | 30 | 25 | 0.564 |
| LMM | 53 | 25 | 28 | |
| Site | ||||
| Sun exposed | 52 | 29 | 23 | 0.344 |
| Sun protected | 56 | 26 | 30 | |
| Stage | ||||
| I/II | 47 | 11 | 36 | <0.001 |
| III/IV | 61 | 44 | 17 | |
LMM=lentigo maligna melanoma; SSM= superficial spreading melanoma
Quantitative reverse transcriptase PCR (qRT-PCR) assay
The expression of miR-126 was determined by qRT-PCR assay. Briefly, total RNA was extracted from the tissues using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. miRNA expression levels were then quantitated using the TaqMan miRNA real-time RT-PCR kit (Applied Biosystems) according to the manufacturer’s protocol. Data were analyzed using 7500 software v.2.0.1 (Applied Biosystems), with the automatic Ct setting for adapting baseline and threshold for Ct determination. The universal small nuclear RNA U6 (RNU6B) was used as an endogenous control for miRNAs. Each sample was examined in triplicate, and the amounts of PCR products produced were non-neoplasticized to RNU6B.
Statistical analysis
Associations between miR-126 expression and various clinicopathological characteristics were analyzed using the χ2 test. Survival times were counted from the date of presentation to the date of death or last follow-up time. Survival rate was determined with Kaplan-Meier and statistically analyzed with the log-rank method between groups. Survival data were evaluated through multivariate Cox regression analysis. P values of <0.05 were considered statistically significant. All statistical analyses were performed using the SPSS 18.0 software (SPSS, Chicago, IL).
Results
Expression level of miR-126 in cutaneous melanoma
qRT-PCR assay was performed to examine the expression of miR-126 in 16 cases of dysplastic nevi, 18 cases of melanoma metastases and 108 cases of primary cutaneous melanoma tissue samples. We found that there were significant differences in the pattern of relative miR-126 expression (shown in Figure 1). Significant differences for miR-126 expression were shown between dysplastic nevi and primary cutaneous melanoma (P<0.01), between primary melanoma and metastatic cutaneous melanomas (P<0.01), and between primary cutaneous melanomas and metastatic cutaneous melanomas (P<0.001).
Figure 1.
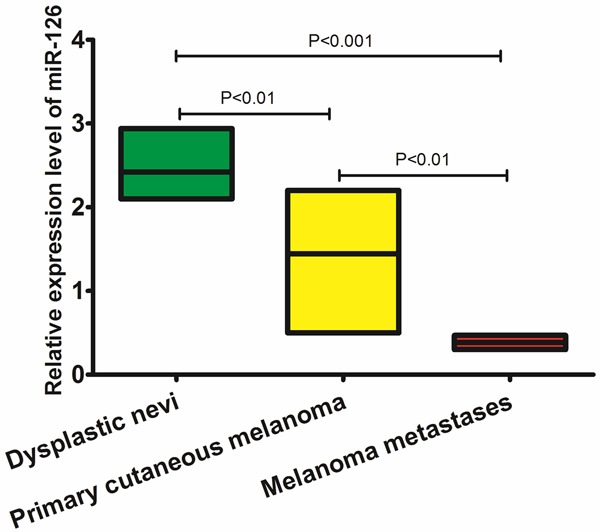
Expression level of miR-126 by qRT-PCR.
Correlation between miR-126 expression and clinicopathological variables of patients with cutaneous melanoma
In this study, patients with values less than the median expression level in tumor tissues were assigned to the low expression group (n=55), whereas those with values more than the median expression level were assigned to the high expression group (n=53). As shown in Table 1, tissue miR-126 expression level was correlated with Breslow thickness (P=0.048), tumor ulceration (P<0.001), and advanced clinical stage (P<0.001). However, tissue miR-126 expression level was not associated with other clinicopathological factors of patients, including age (P=0.422), sex (P=342), histological type (P=0.564), and tumor site (P=0.344).
Prognostic value of miR-126 in cutaneous melanomas
To assess whether the expression of miR-126 was a tumor prognostic biomarker, the overall survival was investigated with respect to expression levels of miR-126 in primary cutaneous melanoma. A total of 108 patients included in the study during the follow-up period and the survival curves plotted by Kaplan-Meier method were shown. As shown in Figure 2, the patients with low miR-126 expression showed shorter 5-year overall survival than those with high miR-126 expression (P=0.039; log-rank test). Table 2 showed the multivariate analysis of the clinicopathological factors related to patient prognosis. Multivariate regression analysis showed that the status of miR-126 expression was an independent prognostic factor overall survival (HR=3.782, 95% CI: 2.479-16.334, P=0.005). Thus, low miR-126 expression was correlated with the poorer overall survival of patients with cutaneous melanoma.
Figure 2.
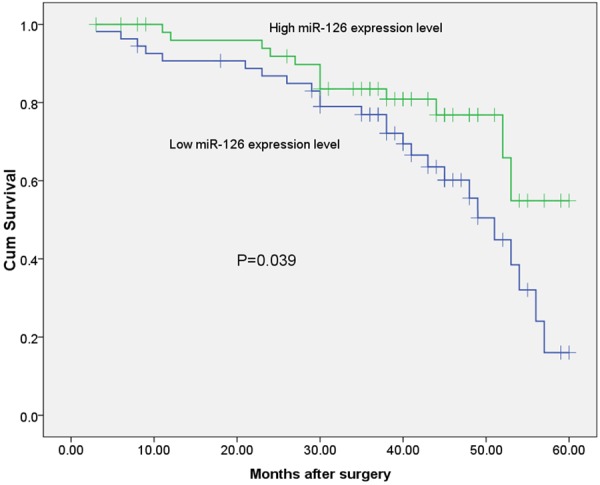
Kaplan-Meier survival curve of overall survival according to miR-126 expression level.
Table 2.
Multivariate analyses of prognostic parameters in 108 patients with primary cutaneous melanomas by Cox regression analysis
| Variable | Hazard ratio | 95% confidence interval | P-value |
|---|---|---|---|
| Age | 1.352 | 0.361-1.821 | 0.427 |
| Gender | 0.782 | 0.231-2.452 | 0.682 |
| Thickness | 2.462 | 1.446-10.01 | 0.021 |
| Ulceration | 2.011 | 0.782-7.254 | 0.089 |
| Histologic type | 1.354 | 0.626-2.669 | 0.627 |
| Site | 0.738 | 0.252-1.891 | 0.513 |
| Stage | 4.559 | 2.235-18.993 | 0.009 |
| miR-126 expression | 3.782 | 2.479-16.334 | 0.005 |
Discussion
Cutaneous melanoma is a common form of cutanous malignancy arising from the pigment cell of the skin, and its incidence is increasing in the US as well as in other parts of the Western world [1,12,13]. Although surgical excision is mostly a definitive treatment at the early stages of the disease, at present standard treatments are ineffective after metastatic dissemination and patients with advanced disease have a severe prognosis [14,15]. Many efforts have been made to develop an understanding of the causes of melanoma progression and more effective therapies. However, they have met with limited success. As melanoma is a highly malignant cancer, an approach that reduces its growth and progression potential may facilitate the development of an effective strategy for its prevention or treatment. In addition, exploring new prognostic markers is conducive to the choice of treatment.
miRNAs have been demonstrated to play functional roles in all the major cellular processes, including tumorigenesis, where they can act as oncogenes as well as tumor suppressor genes, providing a new level of molecular regulation. Researchers are attempting to exploit and identify miRNAs that may serve as either diagnostic or prognostic markers or therapeutic targets in many different tumor types [16,17]. Although the functions of some miRNAs in a variety of human cancers have been identified, limited data are available about the changes of miRNA expression levels and their roles in cutaneous melanoma.
MiR-126 is frequently downregulated in a variety of malignancies and acts as a potential tumor suppressor. Moreover, low expression of miR-126 has been correlated with poor prognosis in several types of cancer. Donnem et al found that miR-126 was a significant negative prognostic factor in patients with lung cancer by multivariate analyses. Furthermore, miR-126 expression correlated significantly with high VEGF-A expression and the co-expression of miR-126 and VEGF-A had a significant prognostic impact [18]. In the study by Liu et al, multivariate analysis indicated that miR-126 was an independent prognostic factor for overall survival in colorectal cancer (CRC) patients, suggesting its clinical significance as a prognostic predictor [19]. Yu et al found that miR-126 was expressed at low levels in cervical cancer. Upregulation of miR-126 inhibited cervical cancer cell proliferation and enhanced the sensitivity to bleomycin (BLM), suggesting miR-126 might represent a novel approach to cervical cancer treatment. The expression level and function of miR-126 have also been investigated in human melanoma. Previously, Felli et al demonstrated that miR-126 played a tumor suppressor role in human melanoma by directly regulating a disintegrin and metalloprotease domain 9 (ADAM9) and metalloprotease 7 (MMP7) [20]. However, the clinical significance of miR-126 in cutaneous melanoma has not been reported.
In the present study, significant differences for miR-126 expression were shown between dysplastic nevi and primary cutaneous melanoma, between primary melanoma and metastatic cutaneous melanomas, and between primary cutaneous melanomas and metastatic cutaneous melanomas. We then found that miR-126 expression level was correlated with Breslow thickness, tumor ulceration, and advanced clinical stage. To assess whether the expression of miR-126 was a tumor prognostic biomarker, the overall survival was investigated with respect to expression levels of miR-126 in primary cutaneous melanoma. The patients with low miR-126 expression showed shorter 5-year overall survival than those with high miR-126 expression. Multivariate regression analysis showed that the status of miR-126 expression was an independent prognostic factor overall survival.
In conclusion, the status of miR-126 might be an independent prognostic factor for patients with cutaneous melanoma. Our results are preliminary and must be verified by large-scale prospective studies.
Disclosure of conflict of interest
None.
References
- 1.Bay C, Kejs AM, Storm HH, Engholm G. Incidence and survival in patients with cutaneous melanoma by morphology, anatomical site and TNM stage: A Danish Population-based Register Study 1989-2011. Cancer Epidemiol. 2015;39:1–7. doi: 10.1016/j.canep.2014.10.010. [DOI] [PubMed] [Google Scholar]
- 2.Erdmann F, Lortet-Tieulent J, Schuz J, Zeeb H, Greinert R, Breitbart EW, Bray F. International trends in the incidence of malignant melanoma 1953-2008--are recent generations at higher or lower risk? International Journal of Cancer Journal International du Cancer. 2013;132:385–400. doi: 10.1002/ijc.27616. [DOI] [PubMed] [Google Scholar]
- 3.Terando A, Sabel MS, Sondak VK. Melanoma: adjuvant therapy and other treatment options. Current Treat Options Oncol. 2003;4:187–199. doi: 10.1007/s11864-003-0020-0. [DOI] [PubMed] [Google Scholar]
- 4.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 5.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fish JE, Srivastava D. MicroRNAs: opening a new vein in angiogenesis research. Sci Signal. 2009;2:pe1. doi: 10.1126/scisignal.252pe1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feng R, Chen X, Yu Y, Su L, Yu B, Li J, Cai Q, Yan M, Liu B, Zhu Z. miR-126 functions as a tumour suppressor in human gastric cancer. Cancer Lett. 2010;298:50–63. doi: 10.1016/j.canlet.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Li XM, Wang AM, Zhang J, Yi H. Down-regulation of miR-126 expression in colorectal cancer and its clinical significance. Med Oncol. 2011;28:1054–1057. doi: 10.1007/s12032-010-9637-6. [DOI] [PubMed] [Google Scholar]
- 9.Negrini M, Calin GA. Breast cancer metastasis: a microRNA story. Breast Cancer Res. 2008;10:203. doi: 10.1186/bcr1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X, Tang S, Le SY, Lu R, Rader JS, Meyers C, Zheng ZM. Aberrant expression of oncogenic and tumor-suppressive microRNAs in cervical cancer is required for cancer cell growth. PLoS One. 2008;3:e2557. doi: 10.1371/journal.pone.0002557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Felli N, Felicetti F, Lustri AM, Errico MC, Bottero L, Cannistraci A, De Feo A, Petrini M, Pedini F, Biffoni M. miR-126&126* restored expressions play a tumor suppressor role by directly regulating ADAM9 and MMP7 in melanoma. PLoS One. 2013;8:e56824. doi: 10.1371/journal.pone.0056824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qureshi AA, Zhang M, Han J. Heterogeneity in host risk factors for incident melanoma and non-melanoma skin cancer in a cohort of US women. J Epidemiol. 2011;21:197–203. doi: 10.2188/jea.JE20100145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gandini S, Sera F, Cattaruzza MS, Pasquini P, Abeni D, Boyle P, Melchi CF. Meta-analysis of risk factors for cutaneous melanoma: I. Common and atypical naevi. Europ J Cancer. 2005;41:28–44. doi: 10.1016/j.ejca.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 14.Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S. Final version of 2009 AJCC melanoma staging and classification. J. Clin. Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ko JM, Fisher DE. A new era: melanoma genetics and therapeutics. The Journal of pathology. 2011;223:241–250. doi: 10.1002/path.2804. [DOI] [PubMed] [Google Scholar]
- 16.Nana-Sinkam SP, Croce CM. Non-coding RNAs in cancer initiation and progression and as novel biomarkers. Molecular Oncol. 2011;5:483–491. doi: 10.1016/j.molonc.2011.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–798. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 18.Donnem T, Lonvik K, Eklo K, Berg T, Sorbye SW, Al-Shibli K, Al-Saad S, Andersen S, Stenvold H, Bremnes RM. Independent and tissue-specific prognostic impact of miR-126 in nonsmall cell lung cancer: coexpression with vascular endothelial growth factor-A predicts poor survival. Cancer. 2011;117:3193–3200. doi: 10.1002/cncr.25907. [DOI] [PubMed] [Google Scholar]
- 19.Liu Y, Zhou Y, Feng X, Yang P, Yang J, An P, Wang H, Ye S, Yu C, He Y, Luo H. Low expression of microRNA-126 is associated with poor prognosis in colorectal cancer. Genes Chromosomes Cancer. 2014;53:358–365. doi: 10.1002/gcc.22146. [DOI] [PubMed] [Google Scholar]
- 20.Yu Q, Liu SL, Wang H, Shi G, Yang P, Chen XL. miR-126 Suppresses the proliferation of cervical cancer cells and alters cell sensitivity to the chemotherapeutic drug bleomycin. APJCP. 2013;14:6569–6572. doi: 10.7314/apjcp.2013.14.11.6569. [DOI] [PubMed] [Google Scholar]


