Fig. 5.
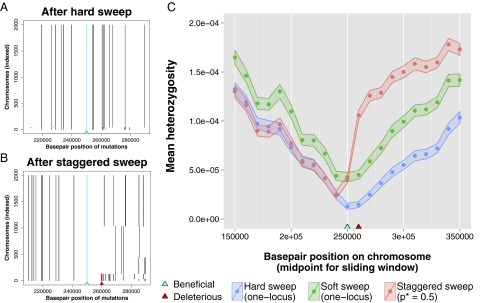
Altered signatures of selection in the genome after a staggered sweep. Simulations were performed using SliM (68) to generate and track neutral diversity around the adaptive site, where simulations used and N = 1,000 diploids. (A) Haplotypes present in a population at the conclusion of single simulation of a hard sweep in which a beneficial mutation on a haplotype containing only neutral mutations was seeded at establishment frequency; note that a single haplotype dominates the population. (B) Haplotypes present in a population at the conclusion of single simulation of a staggered sweep in which a beneficial mutation on a haplotype containing both neutral mutations and a single recessive deleterious mutation () 10 kb away (where ) was seeded at establishment frequency. Note that recombination has unlinked the beneficial and recessive deleterious mutations, such that multiple haplotypes are at high frequency in the population after fixation of the beneficial mutation. (C) Mean heterozygosity across 200 simulations calculated in sliding windows of length 30 kb with step size 10 kb, where the ribbon around data points indicates the SEM. Results are plotted for hard sweep simulations in which a new adaptive mutation occurs on a single haplotype, soft sweep simulations in which a new adaptive mutation occurs on multiple haplotypes (Nub ≈ 1), and staggered sweep simulations in which an adaptive mutation occurs on a single haplotype background containing a recessive deleterious mutation ().
