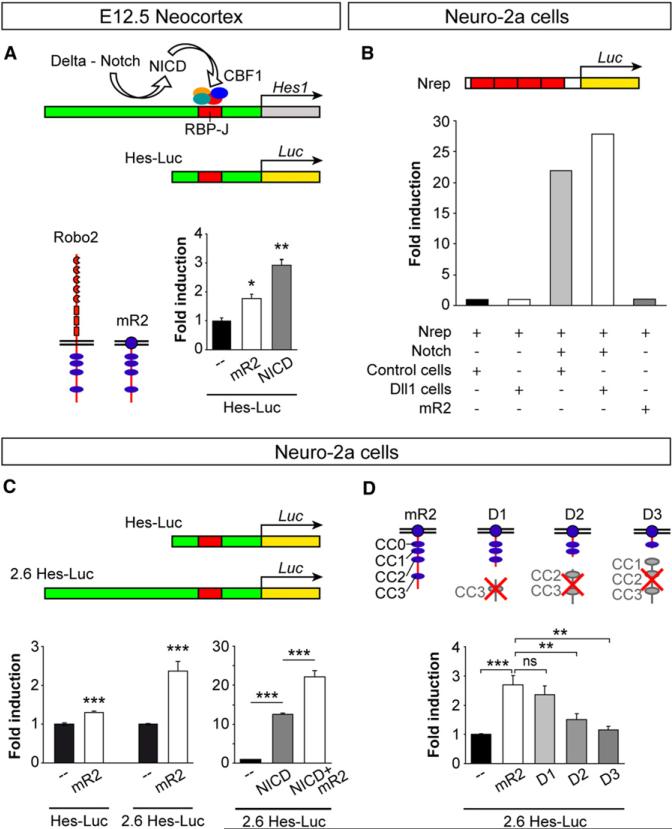
Figure 8. Robo Signaling Drives Hes1 Transcription.
(A) In the developing brain, delta-mediated processing of Notch releases NICD, which interacts with a transcription factor complex that includes CBF1 to activate Hes1 transcription through RBP-J consensus sequences. The Hes-Luc construct tested contains an RBP-J sequence. Schemas depict the basic structure of full-length Robo2 and the myristoylated version of Robo2 (mR2). The histogram shows fold induction of luciferase (Luc) activity from the Hes-Luc construct in E12.5 cortical primary cultures after transfection with mR2 or NICD. Mean ± SEM; t test, *p < 0.05; **p < 0.01.
(B) Hes1 transcription is not activated in Neuro-2a cells upon transstimulation of the Notch pathway with delta. Activation of Hes1 transcription was assayed with a Notch reporter construct (Nrep) containing four RBP-J repeats.
(C) Two different Hes-Luc constructs were used with Neuro-2a cells: Hes-Luc and 2.6 Hes-Luc; the latter includes a long 5 ft region. The graphs show fold induction of Hes-Luc and 2.6 Hes-Luc lucif-erase activities after transfection of Neuro-2a cells with mR2, NICD, or NICD+mR2. Mean ± SEM; t test, ***p < 0.001.
(D) Structure of mR2 and three truncated forms (D1, D2, and D3) and fold induction of 2.6 Hes-Luc luciferase activity in Neuro-2a cells. Statistical significance indicated for mR2 is with respect to basal activity; all others relate to mR2. In addition, D2 values, but not D3, were significantly different than basal values. Mean ± SEM; t test, **p < 0.01; ***p < 0.001.