Figure 3.
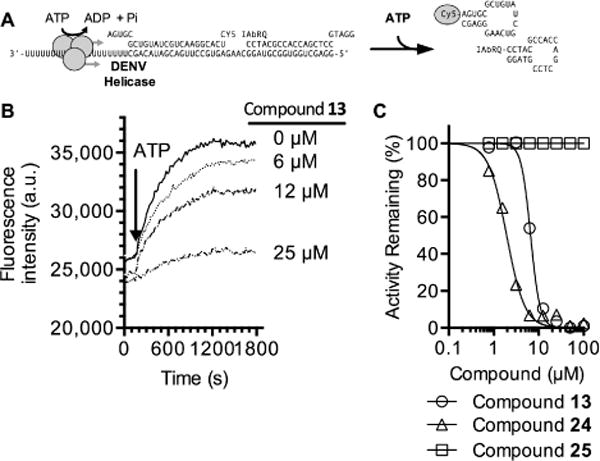
Ability of hit compounds to inhibit DENV helicase-catalyzed RNA strand separation. (A) In the assay, the helicase loads on the 3′ single-stranded tail of the substrate, and translocates 3′ to 5′ upon addition of ATP to separate a Cy5-labeled annealed RNA strand from an annealed DNA strand labeled with the fluorescence quencher Iowa black RQ (IAbRQ, Integrated DNA Technologies).12,37 (B) Concentration-dependent inhibition of DENV helicase-catalyzed RNA unwinding by ML283 (compound 13). Fluorescence traces with only three ML283 concentrations are shown for clarity. (C) Concentration–response plots obtained for select inhibitors. Activity was normalized by first calculating initial rates of fluorescence increase and then normalizing these rates to those observed in negative control reactions containing DMSO alone. IC50 values obtained by fitting data with GraphPad PRISM are shown in Table 1.
