Fig 6. In-frame TAG replacement of KGF.
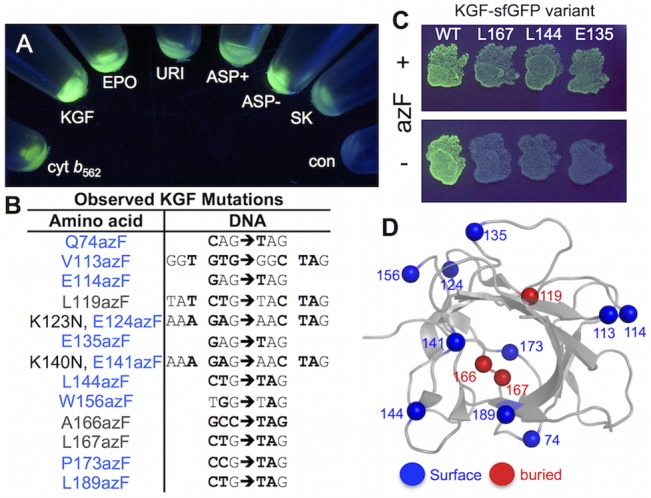
(A) Compatibly of various therapeutic proteins with the E. coli sfGFP fusion screening approach. KGF, keratinocyte growth factor fragment; EPO, ethropoietin; URI, Uricase; ASP+ Asparaginase with N-terminal signal pepetide sequence; ASP- Asparaginase lacking N-terminal signal peptide sequence; SK, Streptokinase. The ‘con’ sample is the control cell sample. The proteins shown are all fused to sfGFP via the TEV digestion sequence. (B) Sequence of observed KGF variants. (C) azF-dependent expression of selected KGF-sfGFP variants. (D) Map of observed mutations on the structure of KGF, including an indication of whether the native residue is surface exposed (blue spheres) or buried (grey spheres) based on surface accessibility.
