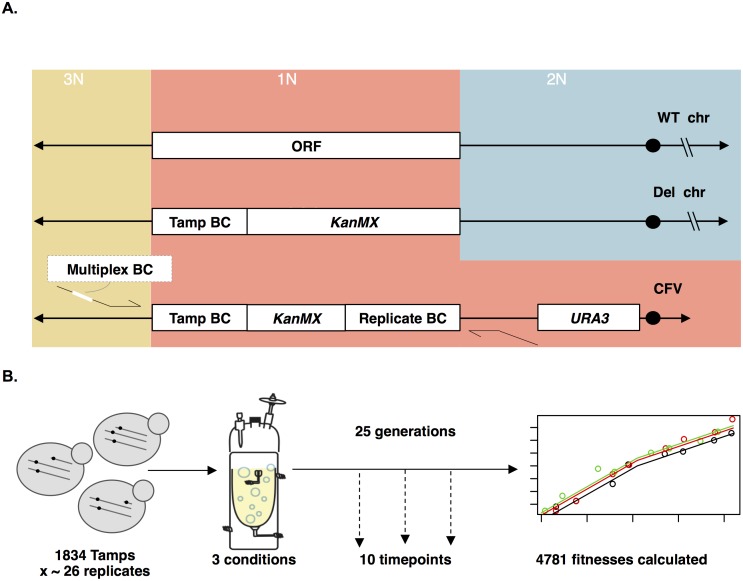
Fig 4. Experimental design for genome-wide screen for the fitness effects of Tamps.
A) A genome-wide pool of telomeric amplicon strains (Tamps) was constructed. Each Tamp initiates at the KanMX cassette and extends to the proximal telomere, creating a strain that has two chromosomal copies (2N) at most genomic locations, one copy (1N) in the region replaced by the KanMX cassette in the deletion collection, and three copies (3N) at locations telomeric of the deleted gene. The Tamp BC and a portion of KanMX are also present at 2 copies. Each strain contains two barcodes: one identifying the Tamp and a second identifying the unique biological replicate. A third barcode was incorporated during the generation of the barcode sequencing libraries which allowed for multiplexing of experimental samples. Large black arrows represent telomeres; large black circles represent centromeres. The primers represent the barseq primers used to create libraries for sequencing. B) Genome-wide pooled competition experimental design.