Abstract
Decoding stem cell metabolism has implicated a tight linkage between energy metabolism and cell fate regulation, a dynamic interplay vital in the execution of developmental and differentiation programs. The inherent plasticity in energy metabolism enables prioritization of metabolic pathways in support of stage-specific demands. Beyond traditional support of energetic needs, intermediate metabolism may also dictate cell fate choices through regulation of cellular signaling and epigenetic regulation of gene expression. The notion of a “metabolism-centric” control of stem cell differentiation has been informed by developmental embryogenesis based upon an on-demand paradigm paramount in defining diverse developmental behaviors, from a post-fertilization nascent zygote to complex organogenesis leading to adequate tissue formation and maturation. Monitored through natural or bioengineered stem cell surrogates, nutrient-responsive metabolites are identified as mediators of cross-talk between metabolic flux, cell signaling, and epigenetic regulation charting, collectively, whether a cell will self-renew to maintain progenitor pools, lineage specify to ensure tissue (re)generation or remain quiescent to curb stress damage. Thus, bioenergetics are increasingly recognized as integral in governing stemness and associated organogenic decisions, paving the way for metabolism-defined targets in control of embryology, stem cell biology and tissue regeneration.
Keywords: Bioenergetics, Differentiation, Embryogenesis, Metabolomics, Stem cells, Regenerative medicine, Reprogramming
From the early zygote to a multicellular organism, evolving demands place distinct metabolic requirements on embryogenesis to provide both sufficient energy and anabolic precursors in support of cellular replication and specialization. Beyond the traditional energy supply role, it is increasingly apparent that metabolic circuits engage genetic programs in order to execute lineage specification algorithms. The interface of metabolism with (epi)genetics is implicated in governing the identity and functional behavior of developing/differentiating cell phenotypes, actuated by nutrient availability and prioritized metabolic pathways. Here, we provide an overview pertinent to the understanding of the metabolic control of developmental and differentiation programs, an emerging paradigm made possible by recent advances in metabolomics technology and development biology platforms.
Metabolic requirements in embryogenesis
The developing embryo transits between stages with rapidly changing anabolic and catabolic demands, which necessitates evolving of the metabolic infrastructure and associated pathways to match these demands. The energy demand imposed by a developing embryo is supplied through the partial breakdown of biomolecules such as carbohydrates and lipids, which in turn drive converging biosynthetic pathways, such as protein and nucleotide biosynthesis required to ensure the success of embryonic development (Figure 1). Oocyte maturation is accompanied by significant mitochondrial biogenesis resulting in a 2–3 order of magnitude increase in mitochondrial content (Piko and Matsumoto 1976; Jansen and Burton 2004), which is thought to occur in preparation to meet the energetic demands of fertilization and early cleavage events during development (Tilly and Sinclair 2013). Although mitochondria of the oocyte and early embryo are characterized by being small with few cristae and electron dense matrices, the mitochondria remain active to engage oxidative ATP generation in support of the metabolic requirements of early development (Figure 2) (Van Blerkom et al. 1995; Motta et al. 2000; Dumollard et al. 2007). Mitochondrial morphology and function is tightly correlated with fertilization and development competence (Santos et al. 2006; Spikings et al. 2007), with mitochondrial DNA (mtDNA) content representing a potential reliable marker of oocyte competence (Tilly and Sinclair 2013). Indeed a minimum threshold of mtDNA appears to be required to enable oocyte progression through meiotic maturation and for successful embryogenesis (Piko and Taylor 1987; Reynier et al. 2001; El Shourbagy et al. 2006; Santos et al. 2006; Wai et al. 2010), consistent with observations that suppressing mtDNA replication during oocyte maturation impairs fertilization and results in arrested embryonic development (Spikings et al. 2007). In addition, abnormal mitochondrial morphology such as disruption of cristae and mitochondrial swelling, which are observed in women in their forties, are correlated with developmental incompetence, strongly supporting the concept that mitochondrial structure and function is a contributor to aging induced decline in oocyte and embryo quality.
Figure 1. Core metabolic principles.
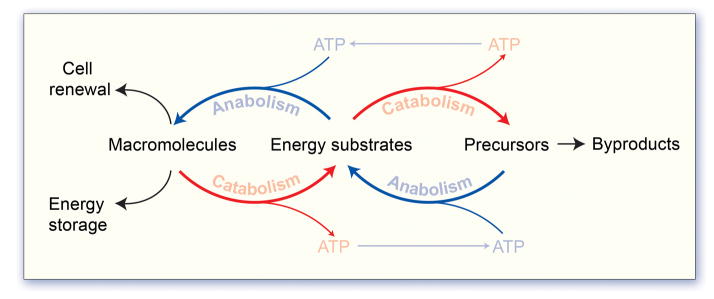
Vital processes are fueled though the metabolism of energy substrates supplied by the environment, such as glucose, fatty acids and amino acids. Catabolism, the process of breaking down (oxidizing) metabolites to produce energy, and anabolism, the process of constructing macromolecules from precursors, are intimately balanced such that catabolic products, including hydrocarbons and energy in the form of ATP and reducing cofactors (NADPH), serve as substrates for the anabolic production of macromolecules that cannot be obtained from the environment. Macromolecules that include lipids, proteins, nucleotides and glycogen represent a cellular energy reserve, and also compose the essential building blocks for cell renewal.
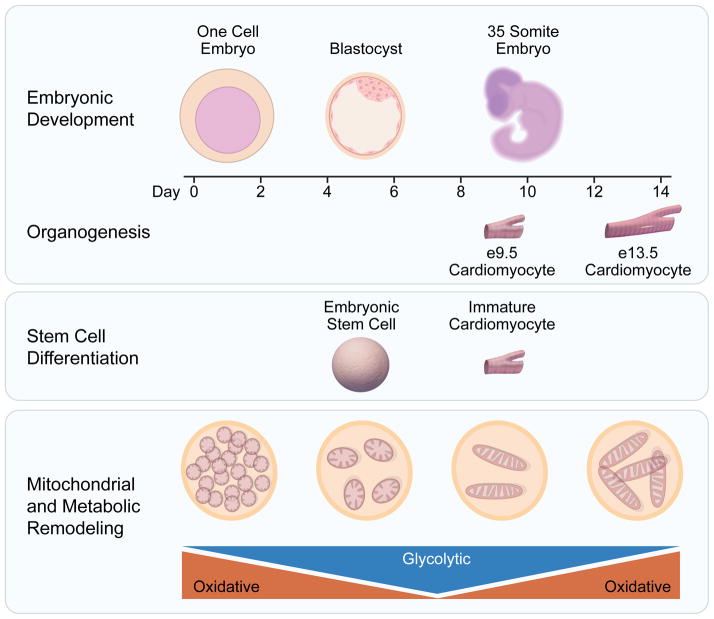
Figure 2. Energy metabolism fuels developmental organogenesis and stem cell differentiation.
Developmental organogenesis reflects an ongoing metamorphosis in energy metabolism, underscoring the ongoing interplay of infrastructural metabolic heritage and evolving bioenergetic needs. Early embryos inherit maternal mitochondria from the oocyte and are largely dependent on oxidative metabolism. Mitochondrial replication is arrested in mature metaphase II eggs and is only reinitiated following transplantation, hence each cell division results in halving of the mitochondrial content of each daughter cell resulting in a decline in oxidative capacity and a greater reliance on glycolysis for energy production. Although less information is available concerning the metabolic maturation of specific organs, evidence indicates that the cardiomyocyte differentiation that occurs between embryonic days 9.5 and 13.5 is dependent upon mitochondrial maturation. Organ specific development can also be model in vitro using stem cell differentiation, which also involves a metabolic transition from predominantly glycolysis to mitochondrial-dependent oxidative metabolism.
Mitochondrial replication is arrested in mature metaphase II eggs and is only reactivated upon implantation into the uterine wall (Larsson et al. 1998), thus the large oocyte mitochondrial pool is segregated across daughter cells during early cell divisions in the pre-implantation embryo. mtDNA is solely maternally inherited as paternal mitochondria are degraded in newly formed embryos, therefore such mitochondrial segregation enables generation of blastomeres with low levels of mitochondria, enabling selection against progeny containing mtDNA mutations that impair metabolic function and prevention of mutational meltdown in subsequent generations (Fan et al. 2008; Shoubridge and Wai 2008). In parallel, after fertilization mitochondria undergo dramatic maturation into the elongated and cristae-rich structures in the blastomeres that more closely resemble ultrastructure in somatic cells (Van Blerkom et al. 1973; Van Blerkom 1989; Van Blerkom 1993; Sathananthan and Trounson 2000). Indeed, failure to obtain this mature mitochondrial infrastructure is often observed in arrested embryos (Van Blerkom 1989), suggesting a critical importance in support of energy generation in the early embryo.
Based upon mitochondrial infrastructure remodeling and changes in the immediate microenvironment, intermediary metabolism also undergoes distinct changes during early embryogenesis. Immediately post-fertilization, the single-cell embryo relies initially on oxidative metabolism, favoring pyruvate generated by the ovarian follicle cells as a primary metabolic substrate owing to inheritance of abundant maternal mitochondria within the oocyte. Glycolytic rates are initially low due to inhibition of hexokinase and phosphofructose kinase 1, the rate limiting steps of glycolysis (Barbehenn et al. 1978). Indeed, high levels of glucose can actually impair normal early embryo development (Brinster and Troike 1979). Inhibition of mitochondrial oxidative metabolism impairs oocyte maturation, fertilization and embryonic development implicating provision of sufficient ATP as a critical determinant of embryogenesis (Van Blerkom et al. 1995), as lack of ATP generation being linked to defective chromosomal segregation and spindle abnormalities (Schon et al. 2000; Eichenlaub-Ritter et al. 2004; Zheng et al. 2007). As mitochondria content per cell declines during morula compaction and subsequent blastomere formation, glucose transporter expression increases such that glucose uptake gradually accelerates until it exceeds that of pyruvate or lactate (Pantaleon and Kaye 1998). Due to increased glucose availability, glycolysis is accelerated in the blastocyst, whereby all glucose is metabolized to lactate, which may occur in preparation for implantation into the hypoxic uterine wall. The importance of glycolysis is underscored as mutations in several glycolytic enzymes results in early post-implantation lethality (Merkle and Pretsch 1989; West et al. 1990; Merkle et al. 1992; Pretsch 2000; Johnson et al. 2003). Following implantation, as blood flow is established to the embryo, mitochondrial biogenesis and maturation engages to support the re-establishment of oxidative metabolism leading to the progressive decline in glycolysis, consistent with the developmental delay or late onset lethality with mutations that impair oxidative metabolism or mitochondria biogenesis (Johnson et al. 2003; Van Blerkom 2009).
Although the transition in mitochondria and energy metabolism is well characterized in early embryogenesis, much less is know how mitochondrial maturation contributes to the development of specific lineages. Initial evidence indicates that mitochondrial maturation is also critical for cardiomyocyte development, with early cardiomyocytes displaying immature fragmented mitochondria, which develop into extensive networks of filamentous mitochondria by day 13.5 of mouse development (Hom et al. 2011). Cardiomyocyte maturation is dependent on this mitochondrial development as arresting mitochondrial maturation impairs cardiomyocyte differentiation, while differentiation can be accelerated by promoting mitochondrial maturation (Hom et al. 2011; Folmes et al. 2012b). However, the impact of this on development of cardiac specific metabolic processes remains unexamined, although switching of energy metabolism from glycolysis to oxidative metabolism is critical for the developing retina (Agathocleous et al. 2012), suggesting that a similar process is occurring during cardiac development. Taken together, metabolic fluidity in energetic substrate choices is a critical contributor to normal development.
Metabolism supported stemness
Establishment of stem cell platforms has enabled the recapitulation of embryonic development in vitro and has facilitated the decoding of metabolic processes supporting cell fate decisions. Embryonic stem cells derived from the inner cell mass of the blastocyst, represent the gold-standard of stemness, namely displaying the defining features of “pluripotency”, the aptitude to differentiate into diverse tissue-specific lineages, and “self-renewal”, the ability to self-replicate while in the undifferentiated state. Like their tissue of origin, embryonic stem cells have a low mtDNA copy number and contain immature, spherical and cristae poor mitochondria (Chung et al. 2007)., which is a consistent feature across a number of stem cell populations (Piccoli et al. 2005; Lonergan et al. 2006; Lonergan et al. 2007; Chen et al. 2008; Folmes et al. 2011b). This infrastructure is associated with reduced oxidative capacity, with embryonic stem cells respiring near their maximal rates, even under basal unstimulated conditions (Cho et al. 2006; Chung et al. 2007; Kondoh et al. 2007; Chen et al. 2008; Turner et al. 2008; Simsek et al. 2010; Folmes et al. 2011a). Despite the apparent immaturity of stem cell mitochondria, disruption of mitochondrial homeostatis results in loss of pluripotency and impaired differentiation capacity (Facucho-Oliveira et al. 2007; Todd et al. 2010), indicating that stem cells may repurpose mitochondria from energy generators to alternative functions in support of stemness maintenance. Stem cells are typically dependent on glycolysis to provide not only sufficient energy to maintain cell homeostasis, but also to supply carbon biomass for the biosynthesis of cellular building block including nucleotides, proteins and lipids to support proliferation and self renewal. Indeed, inhibition of glycolysis impairs stem cell differentiation and induces cell death, while methods that promote glycolysis facilitates stemness maintenance (Ezashi et al. 2005; Kondoh et al. 2007; Varum et al. 2009; Mohyeldin et al. 2010; Chen et al. 2011).
The bioenergetics requirements of pluripotency are underscored by the metabolic transitions that occur during embryo-independent derivation of stem cell through nuclear reprogramming. Coerced expression of stemness transcription factors into somatic cells resets the pluripotent ground state, and enables generation of induced pluripotent stem (iPS) cell lines capable to self-renew and to yield lineage specialized progeny (Takahashi and Yamanaka 2006). Bioengineered stem cell populations recapitulate features of natural pluripotent counterparts through structural and functional metamorphosis, supported by coordinated metabolic reprogramming (Folmes et al. 2012a). Indeed, longitudinal proteomic profiling of nuclear reprogramming indicates that remodeling of the mitochondrial electron transport chain is among the earliest events on route to pluripotency (Hansson et al. 2012). Nuclear reprogramming imposes massive mitochondrial restructuring with transition from the mature cristae-rich mitochondria defining the somatic phenotype to immature cristae-poor mitochondria with reduced mitochondrial DNA copy number characteristic of derived iPS cells (Prigione et al. 2010; Folmes et al. 2011a; Varum et al. 2011). However, pluripotent cells maintain a high mitochondrial membrane potential, which would permit conservation of an energetically primed state able to respond to increase in energetic demands, such as that imposed by differentiation. The mitochondrial remodeling results in a suppression of cellular respiration in iPS cells, supporting the switch from somatic oxidative phosphorylation in favor of glycolysis that characterizes nuclear reprogramming (Folmes et al. 2011a; Panopoulos et al. 2012). Indeed, higher reprogramming efficiency is observed upon stimulation of glycolysis (Yoshida et al. 2009; Zhu et al. 2010; Folmes et al. 2011a) or by utilizing somatic sources with a more glycolytic metabolic phenotype (Panopoulos et al. 2012), while reprogramming is attenuated by inhibiting glycolysis or promoting oxidative metabolism (Zhu et al. 2010; Folmes et al. 2011a).
Beyond the balance between glycolysis and oxidative metabolism, pluripotent stem cells also display a high requirement for catabolism of specific amino acids. Mouse pluripotent stem cells express high levels of threonine dehydrogenase, which catalyzes the first step of conversion of the amino acid threonine into glycine and acetyl-CoA (Wang et al. 2009; Shyh-Chang et al. 2013). Removal of threonine from cell culture impairs stem cell proliferation, while inhibition of threonine dehydrogenase disrupts stem cell colony growth and impairs the nuclear reprogramming (Wang et al. 2009; Alexander et al. 2011; Han et al. 2013; Shyh-Chang et al. 2013). Glycine produced from threonine catabolism is further catabolized by glycine decarboxylase to provide 1-carbon equivalents for the folate pool, which ultimately supports the S-adenosylmethionine pathway and regulation of histone H3K4 methylation, which is critical for pluripotent stem cell function (Shyh-Chang et al. 2013). Threonine dehydrogenase is only expressed as a nonfunction pseudogene in humans, however it appears that high flux through methionine metabolism plays a similar role in maintaining SAM pathway intermediates (Shiraki et al. 2014).
Meeting the needs of differentiated progeny
Differentiated cells have high energetic demands in line with specialized tissue functions. These demands are typically met by complete oxidation of metabolic substrates including carbohydrates, fatty acids and amino acids in the mitochondrial tricarboxylic acid cycle to support maximum generation of ATP. Accordingly, differentiation of pluripotent stem cells into lineage-specified progeny requires remodeling of bioenergetic systems to support the catabolic generation of ATP at the expense of glycolysis. Upon downregulation of pluripotent genes, mtDNA replication and mitochondrial biogenesis accelerates, concomitant with upregulation of tricarboxylic acid enzyme and electron transport chain subunits to support cellular respiration and efficient ATP generation (Chung et al. 2007; Facucho-Oliveira et al. 2007; Tormos et al. 2011). Maturing mitochondria then migrate across cytoplasmic areas to form mitochondrial networks and allow energetic transfers to cytosolic organelles (Cho et al. 2006; Lonergan et al. 2006; Chung et al. 2007; Facucho-Oliveira et al. 2007; Chen et al. 2008). The transition between glycolysis and oxidative metabolism is critical for lineage specification as inhibition of glycolysis or the pentose phosphate pathway promotes myogenic differentiation, while disruption of mitochondrial respiratory chain function compromises the ability of pluripotent stem cells to differentiate into specialized tissues (Chung et al. 2007; Bracha et al. 2010; Mandal et al. 2011). Differentiation also engages a maturation of the metabolic signaling and phosphotransfer infrastructure, including increased expression of AMP-activated protein kinase, creatine kinase and adenlyate kinase (Chung et al. 2008; Chung et al. 2010; Dzeja et al. 2011). These observations suggest that the glycolytic phenotype of pluripotent stem cells is a critical component required to maintain these cells in the pluripotent state, while mitochondrial and metabolic infrastructure maturation may be integral to the differentiation process.
The balance between glycolysis and oxidative metabolism has been established as a rheostat for stem cell fate, however the specific metabolic pathways that support stem cell renewal versus lineage specification are only starting to be examined. For instance, hematopoietic stem cells utilize PPARδ-induced fatty acid oxidation to support asymmetrical cell division producing one daughter cell that retains the stem cell fate, while the other can go on to a differentiated state, in order to ensure stem cell pool maintenance (Ito et al. 2012). In contrast, inhibition of fatty acid oxidation promotes symmetric cell division into stem cell committee daughter cells leading to initial amplification of the progenitor pool, followed by exhaustion (Ito et al. 2012). This is complemented by the requirement for fatty acid synthesis in proliferating neural stem and progenitor cells (Knobloch et al. 2013), indicating that the regulation of fatty acid oxidation versus synthesis may represent a critical component of stem cell fate regulation (Folmes et al. 2013). Although elevation of oxidative metabolism appears to be a consistent feature of lineage specification, it remains largely unexamined how specific metabolic pathways may drive cell along discrete lineages. Indeed a recent study has demonstrated that utilization of glutaminolysis and nucleotide biosynthesis versus glucose catabolism represents a branch point between erythroid and myeloid fates during hematopoietic stem cell lineage specification (Oburoglu et al. 2014). Of particular interest is that glutaminolysis is only critical for early erythroid commitment and is not necessary for maintenance of this lineage, thus implicating that a specific metabolic state is required to prime and initiate a distinct lineage-specification and not simply match energy metabolism to the demands of that lineage (Folmes and Terzic 2014).
Summary
The metabolic determinants of early embryogenesis have been broadly established, requiring metabolic fluidity to meet discrete anabolic and catabolic demands of developmental stages. Yet, how metabolism changes during tissue-specific development is less known. Recent advances in stem cell biology and the refinement of metabolic profiling techniques have increasingly enabled the dissection of the metabolic contribution to cell fate. Particular emphasis has been placed on defining the requirements associated with the maintenance of stem cell pluripotency and self-renewal, as well as on specification into defined lineages. While it has been documented that glycolysis is essential in stemness and oxidative phosphorylation in support of differentiated states, individual pathways that are critical for commitment of specific cell fate decisions have started to be identified. Establishing a comprehensive map of cellular bioenergetics across developmental stages and tissue lineages would offer a metabolic foundation underlying development, reproduction and regeneration.
References
- Agathocleous M, Love NK, Randlett O, Harris JJ, Liu J, Murray AJ, Harris WA. Metabolic differentiation in the embryonic retina. Nature Cell Biology. 2012;14(8):859–64. doi: 10.1038/ncb2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander PB, Wang J, McKnight SL. Targeted killing of a mammalian cell based upon its specialized metabolic state. Proc Natl Acad Sci U S A. 2011;108(38):15828–33. doi: 10.1073/pnas.1111312108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbehenn EK, Wales RG, Lowry OH. Measurement of metabolites in single preimplantation embryos; a new means to study metabolic control in early embryos. J Embryol Exp Morphol. 1978;43:29–46. [PubMed] [Google Scholar]
- Bracha AL, Ramanathan A, Huang S, Ingber DE, Schreiber SL. Carbon metabolism-mediated myogenic differentiation. Nature Chemical Biology. 2010;6(3):202–204. doi: 10.1038/nchembio.301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brinster RL, Troike DE. Requirements for blastocyst development in vitro. J Anim Sci. 1979;49(Suppl 2):26–34. doi: 10.1093/ansci/49.supplement_ii.26. [DOI] [PubMed] [Google Scholar]
- Chen CT, Shih YR, Kuo TK, Lee OK, Wei YH. Coordinated changes of mitochondrial biogenesis and antioxidant enzymes during osteogenic differentiation of human mesenchymal stem cells. Stem Cells. 2008;26(4):960–8. doi: 10.1634/stemcells.2007-0509. [DOI] [PubMed] [Google Scholar]
- Chen G, Gulbranson DR, Hou Z, Bolin JM, Ruotti V, Probasco MD, Smuga-Otto K, Howden SE, Diol NR, Propson NE, Wagner R, Lee GO, Antosiewicz-Bourget J, Teng JM, Thomson JA. Chemically defined conditions for human iPSC derivation and culture. Nature Methods. 2011;8(5):424–9. doi: 10.1038/nmeth.1593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho YM, Kwon S, Pak YK, Seol HW, Choi YM, Park do J, Park KS, Lee HK. Dynamic changes in mitochondrial biogenesis and antioxidant enzymes during the spontaneous differentiation of human embryonic stem cells. Biochem Biophys Res Commun. 2006;348(4):1472–8. doi: 10.1016/j.bbrc.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Chung S, Arrell DK, Faustino RS, Terzic A, Dzeja PP. Glycolytic network restructuring integral to the energetics of embryonic stem cell cardiac differentiation. J Mol Cell Cardiol. 2010;48(4):725–34. doi: 10.1016/j.yjmcc.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Perez-Terzic C, Behfar A, Terzic A. Mitochondrial oxidative metabolism is required for the cardiac differentiation of stem cells. Nat Clin Pract Cardiovasc Med. 2007;4(Suppl 1):S60–7. doi: 10.1038/ncpcardio0766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung S, Dzeja PP, Faustino RS, Terzic A. Developmental restructuring of the creatine kinase system integrates mitochondrial energetics with stem cell cardiogenesis. Ann N Y Acad Sci. 2008;1147:254–63. doi: 10.1196/annals.1427.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol. 2007;77:21–49. doi: 10.1016/S0070-2153(06)77002-8. [DOI] [PubMed] [Google Scholar]
- Dzeja PP, Chung S, Faustino RS, Behfar A, Terzic A. Developmental enhancement of adenylate kinase-AMPK metabolic signaling axis supports stem cell cardiac differentiation. PLoS One. 2011;6(4):e19300. doi: 10.1371/journal.pone.0019300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenlaub-Ritter U, Vogt E, Yin H, Gosden R. Spindles, mitochondria and redox potential in ageing oocytes. Reprod Biomed Online. 2004;8(1):45–58. doi: 10.1016/s1472-6483(10)60497-x. [DOI] [PubMed] [Google Scholar]
- El Shourbagy SH, Spikings EC, Freitas M, St John JC. Mitochondria directly influence fertilisation outcome in the pig. Reproduction. 2006;131(2):233–45. doi: 10.1530/rep.1.00551. [DOI] [PubMed] [Google Scholar]
- Ezashi T, Das P, Roberts RM. Low O2 tensions and the prevention of differentiation of hES cells. Proc Natl Acad Sci USA. 2005;102(13):4783–8. doi: 10.1073/pnas.0501283102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Facucho-Oliveira JM, Alderson J, Spikings EC, Egginton S, St John JC. Mitochondrial DNA replication during differentiation of murine embryonic stem cells. J Cell Sci. 2007;120(Pt 22):4025–34. doi: 10.1242/jcs.016972. [DOI] [PubMed] [Google Scholar]
- Fan W, Waymire KG, Narula N, Li P, Rocher C, Coskun PE, Vannan MA, Narula J, Macgregor GR, Wallace DC. A mouse model of mitochondrial disease reveals germline selection against severe mtDNA mutations. Science. 2008;319(5865):958–62. doi: 10.1126/science.1147786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Dzeja PP, Nelson TJ, Terzic A. Metabolic plasticity in stem cell homeostasis and differentiation. Cell Stem Cell. 2012a;11(5):596–606. doi: 10.1016/j.stem.2012.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Martinez-Fernandez A, Arrell DK, Lindor JZ, Dzeja PP, Ikeda Y, Perez-Terzic C, Terzic A. Somatic oxidative bioenergetics transitions into pluripotency-dependent glycolysis to facilitate nuclear reprogramming. Cell Metabolism. 2011a;14(2):264–71. doi: 10.1016/j.cmet.2011.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Nelson TJ, Terzic A. Energy metabolism in nuclear reprogramming. Biomark Med. 2011b;5(6):715–29. doi: 10.2217/bmm.11.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CD, Park S, Terzic A. Lipid metabolism greases the stem cell engine. Cell Metab. 2013;17(2):153–5. doi: 10.1016/j.cmet.2013.01.010. [DOI] [PubMed] [Google Scholar]
- Folmes CD, Terzic A. Stem cell lineage specification: you become what you eat. Cell Metab. 2014;20(3):389–91. doi: 10.1016/j.cmet.2014.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Folmes CDL, Dzeja PP, Nelson TJ, Terzic A. Mitochondria in control of cell fate. Circulation Research. 2012b;110(4):526–529. doi: 10.1161/RES.0b013e31824ae5c1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Gu H, Wang J, Lu W, Mei Y, Wu M. Regulation of L-threonine dehydrogenase in somatic cell reprogramming. Stem Cells. 2013;31(5):953–65. doi: 10.1002/stem.1335. [DOI] [PubMed] [Google Scholar]
- Hansson J, Rafiee MR, Reiland S, Polo JM, Gehring J, Okawa S, Huber W, Hochedlinger K, Krijgsveld J. Highly coordinated proteome dynamics during reprogramming of somatic cells to pluripotency. Cell Rep. 2012;2(6):1579–92. doi: 10.1016/j.celrep.2012.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hom JR, Quintanilla RA, Hoffman DL, de Mesy Bentley KL, Molkentin JD, Sheu SS, Porter GA., Jr The permeability transition pore controls cardiac mitochondrial maturation and myocyte differentiation. Developmental Cell. 2011;21(3):469–78. doi: 10.1016/j.devcel.2011.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito K, Carracedo A, Weiss D, Arai F, Ala U, Avigan DE, Schafer ZT, Evans RM, Suda T, Lee CH, Pandolfi PP. A PML-PPAR-delta pathway for fatty acid oxidation regulates hematopoietic stem cell maintenance. Nat Med. 2012;18(9):1350–8. doi: 10.1038/nm.2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen RP, Burton GJ. Mitochondrial dysfunction in reproduction. Mitochondrion. 2004;4(5–6):577–600. doi: 10.1016/j.mito.2004.07.038. [DOI] [PubMed] [Google Scholar]
- Johnson MT, Mahmood S, Patel MS. Intermediary metabolism and energetics during murine early embryogenesis. Journal of Biological Chemistry. 2003;278(34):31457–60. doi: 10.1074/jbc.R300002200. [DOI] [PubMed] [Google Scholar]
- Knobloch M, Braun SM, Zurkirchen L, von Schoultz C, Zamboni N, Arauzo-Bravo MJ, Kovacs WJ, Karalay O, Suter U, Machado RA, Roccio M, Lutolf MP, Semenkovich CF, Jessberger S. Metabolic control of adult neural stem cell activity by Fasn-dependent lipogenesis. Nature. 2013;493(7431):226–30. doi: 10.1038/nature11689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kondoh H, Lleonart ME, Nakashima Y, Yokode M, Tanaka M, Bernard D, Gil J, Beach D. A high glycolytic flux supports the proliferative potential of murine embryonic stem cells. Antioxid Redox Signal. 2007;9(3):293–9. doi: 10.1089/ars.2006.1467. [DOI] [PubMed] [Google Scholar]
- Larsson NG, Wang J, Wilhelmsson H, Oldfors A, Rustin P, Lewandoski M, Barsh GS, Clayton DA. Mitochondrial transcription factor A is necessary for mtDNA maintenance and embryogenesis in mice. Nature Genetics. 1998;18(3):231–6. doi: 10.1038/ng0398-231. [DOI] [PubMed] [Google Scholar]
- Lonergan T, Bavister B, Brenner C. Mitochondria in stem cells. Mitochondrion. 2007;7(5):289–96. doi: 10.1016/j.mito.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lonergan T, Brenner C, Bavister B. Differentiation-related changes in mitochondrial properties as indicators of stem cell competence. J Cell Physiol. 2006;208(1):149–53. doi: 10.1002/jcp.20641. [DOI] [PubMed] [Google Scholar]
- Mandal S, Lindgren AG, Srivastava AS, Clark AT, Banerjee U. Mitochondrial function controls proliferation and early differentiation potential of embryonic stem cells. Stem Cells. 2011;29(3):486–95. doi: 10.1002/stem.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle S, Favor J, Graw J, Hornhardt S, Pretsch W. Hereditary lactate dehydrogenase A-subunit deficiency as cause of early postimplantation death of homozygotes in Mus musculus. Genetics. 1992;131(2):413–21. doi: 10.1093/genetics/131.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkle S, Pretsch W. Characterization of triosephosphate isomerase mutants with reduced enzyme activity in Mus musculus. Genetics. 1989;123(4):837–44. doi: 10.1093/genetics/123.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150–61. doi: 10.1016/j.stem.2010.07.007. [DOI] [PubMed] [Google Scholar]
- Motta PM, Nottola SA, Makabe S, Heyn R. Mitochondrial morphology in human fetal and adult female germ cells. Hum Reprod. 2000;15(Suppl 2):129–47. doi: 10.1093/humrep/15.suppl_2.129. [DOI] [PubMed] [Google Scholar]
- Oburoglu L, Tardito S, Fritz V, de Barros SC, Merida P, Craveiro M, Mamede J, Cretenet G, Mongellaz C, An X, Klysz D, Touhami J, Boyer-Clavel M, Battini JL, Dardalhon V, Zimmermann VS, Mohandas N, Gottlieb E, Sitbon M, Kinet S, Taylor N. Glucose and glutamine metabolism regulate human hematopoietic stem cell lineage specification. Cell Stem Cell. 2014;15(2):169–84. doi: 10.1016/j.stem.2014.06.002. [DOI] [PubMed] [Google Scholar]
- Panopoulos AD, Yanes O, Ruiz S, Kida YS, Diep D, Tautenhahn R, Herrerias A, Batchelder EM, Plongthongkum N, Lutz M, Berggren WT, Zhang K, Evans RM, Siuzdak G, Belmonte JC. The metabolome of induced pluripotent stem cells reveals metabolic changes occurring in somatic cell reprogramming. Cell Research. 2012;22(1):168–77. doi: 10.1038/cr.2011.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pantaleon M, Kaye PL. Glucose transporters in preimplantation development. Rev Reprod. 1998;3(2):77–81. doi: 10.1530/ror.0.0030077. [DOI] [PubMed] [Google Scholar]
- Piccoli C, Ria R, Scrima R, Cela O, D’Aprile A, Boffoli D, Falzetti F, Tabilio A, Capitanio N. Characterization of mitochondrial and extra-mitochondrial oxygen consuming reactions in human hematopoietic stem cells. Novel evidence of the occurrence of NAD(P)H oxidase activity. J Biol Chem. 2005;280(28):26467–76. doi: 10.1074/jbc.M500047200. [DOI] [PubMed] [Google Scholar]
- Piko L, Matsumoto L. Number of mitochondria and some properties of mitochondrial DNA in the mouse egg. Dev Biol. 1976;49(1):1–10. doi: 10.1016/0012-1606(76)90253-0. [DOI] [PubMed] [Google Scholar]
- Piko L, Taylor KD. Amounts of mitochondrial DNA and abundance of some mitochondrial gene transcripts in early mouse embryos. Dev Biol. 1987;123(2):364–74. doi: 10.1016/0012-1606(87)90395-2. [DOI] [PubMed] [Google Scholar]
- Pretsch W. Enzyme-activity mutants in Mus musculus. I. Phenotypic description and genetic characterization of ethylnitrosourea-induced mutations. Mamm Genome. 2000;11(7):537–42. doi: 10.1007/s003350010103. [DOI] [PubMed] [Google Scholar]
- Prigione A, Fauler B, Lurz R, Lehrach H, Adjaye J. The senescence-related mitochondrial/oxidative stress pathway is repressed in human induced pluripotent stem cells. Stem Cells. 2010;28(4):721–33. doi: 10.1002/stem.404. [DOI] [PubMed] [Google Scholar]
- Reynier P, May-Panloup P, Chretien MF, Morgan CJ, Jean M, Savagner F, Barriere P, Malthiery Y. Mitochondrial DNA content affects the fertilizability of human oocytes. Mol Hum Reprod. 2001;7(5):425–9. doi: 10.1093/molehr/7.5.425. [DOI] [PubMed] [Google Scholar]
- Santos TA, El Shourbagy S, St John JC. Mitochondrial content reflects oocyte variability and fertilization outcome. Fertil Steril. 2006;85(3):584–91. doi: 10.1016/j.fertnstert.2005.09.017. [DOI] [PubMed] [Google Scholar]
- Sathananthan AH, Trounson AO. Mitochondrial morphology during preimplantational human embryogenesis. Hum Reprod. 2000;15(Suppl 2):148–59. doi: 10.1093/humrep/15.suppl_2.148. [DOI] [PubMed] [Google Scholar]
- Schon EA, Kim SH, Ferreira JC, Magalhaes P, Grace M, Warburton D, Gross SJ. Chromosomal non-disjunction in human oocytes: is there a mitochondrial connection? Hum Reprod. 2000;15(Suppl 2):160–72. doi: 10.1093/humrep/15.suppl_2.160. [DOI] [PubMed] [Google Scholar]
- Shiraki N, Shiraki Y, Tsuyama T, Obata F, Miura M, Nagae G, Aburatani H, Kume K, Endo F, Kume S. Methionine metabolism regulates maintenance and differentiation of human pluripotent stem cells. Cell Metab. 2014;19(5):780–94. doi: 10.1016/j.cmet.2014.03.017. [DOI] [PubMed] [Google Scholar]
- Shoubridge EA, Wai T. Sidestepping mutational meltdown. Science. 2008;319(5865):914–5. doi: 10.1126/science.1154515. [DOI] [PubMed] [Google Scholar]
- Shyh-Chang N, Locasale JW, Lyssiotis CA, Zheng Y, Teo RY, Ratanasirintrawoot S, Zhang J, Onder T, Unternaehrer JJ, Zhu H, Asara JM, Daley GQ, Cantley LC. Influence of threonine metabolism on S-adenosylmethionine and histone methylation. Science. 2013;339(6116):222–6. doi: 10.1126/science.1226603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simsek T, Kocabas F, Zheng J, Deberardinis RJ, Mahmoud AI, Olson EN, Schneider JW, Zhang CC, Sadek HA. The distinct metabolic profile of hematopoietic stem cells reflects their location in a hypoxic niche. Cell Stem Cell. 2010;7(3):380–390. doi: 10.1016/j.stem.2010.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spikings EC, Alderson J, St John JC. Regulated mitochondrial DNA replication during oocyte maturation is essential for successful porcine embryonic development. Biol Reprod. 2007;76(2):327–35. doi: 10.1095/biolreprod.106.054536. [DOI] [PubMed] [Google Scholar]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126(4):663–76. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- Tilly JL, Sinclair DA. Germline energetics, aging, and female infertility. Cell Metab. 2013;17(6):838–50. doi: 10.1016/j.cmet.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Todd LR, Damin MN, Gomathinayagam R, Horn SR, Means AR, Sankar U. Growth factor erv1-like modulates Drp1 to preserve mitochondrial dynamics and function in mouse embryonic stem cells. Mol Biol Cell. 2010;21(7):1225–36. doi: 10.1091/mbc.E09-11-0937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tormos KV, Anso E, Hamanaka RB, Eisenbart J, Joseph J, Kalyanaraman B, Chandel NS. Mitochondrial complex III ROS regulate adipocyte differentiation. Cell Metabolism. 2011;14(4):537–44. doi: 10.1016/j.cmet.2011.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner WS, Seagle C, Galanko JA, Favorov O, Prestwich GD, Macdonald JM, Reid LM. Nuclear magnetic resonance metabolomic footprinting of human hepatic stem cells and hepatoblasts cultured in hyaluronan-matrix hydrogels. Stem Cells. 2008;26(6):1547–55. doi: 10.1634/stemcells.2007-0863. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Morphodynamics of nuclear and cytoplasmic reorganization during the resumption of arrested meiosis in the mouse oocyte. Prog Clin Biol Res. 1989;294:33–51. [PubMed] [Google Scholar]
- Van Blerkom J. Development of human embryos to the hatched blastocyst stage in the presence or absence of a monolayer of Vero cells. Hum Reprod. 1993;8(9):1525–39. doi: 10.1093/oxfordjournals.humrep.a138293. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J. Mitochondria in early mammalian development. Seminars in Cell and Developmental Biology. 2009;20(3):354–64. doi: 10.1016/j.semcdb.2008.12.005. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Davis PW, Lee J. ATP content of human oocytes and developmental potential and outcome after in-vitro fertilization and embryo transfer. Hum Reprod. 1995;10(2):415–24. doi: 10.1093/oxfordjournals.humrep.a135954. [DOI] [PubMed] [Google Scholar]
- Van Blerkom J, Manes C, Daniel JC., Jr Development of preimplantation rabbit embryos in vivo and in vitro. I. An ultrastructural comparison. Dev Biol. 1973;35(2):262–82. doi: 10.1016/0012-1606(73)90023-7. [DOI] [PubMed] [Google Scholar]
- Varum S, Momcilovic O, Castro C, Ben-Yehudah A, Ramalho-Santos J, Navara CS. Enhancement of human embryonic stem cell pluripotency through inhibition of the mitochondrial respiratory chain. Stem Cell Res. 2009;3(2–3):142–56. doi: 10.1016/j.scr.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varum S, Rodrigues AS, Moura MB, Momcilovic O, Easley CA, Ramalho-Santos J, Van Houten B, Schatten G. Energy metabolism in human pluripotent stem cells and their differentiated counterparts. PLoS One. 2011;6(6):e20914. doi: 10.1371/journal.pone.0020914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wai T, Ao A, Zhang X, Cyr D, Dufort D, Shoubridge EA. The role of mitochondrial DNA copy number in mammalian fertility. Biol Reprod. 2010;83(1):52–62. doi: 10.1095/biolreprod.109.080887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Alexander P, Wu L, Hammer R, Cleaver O, McKnight SL. Dependence of mouse embryonic stem cells on threonine catabolism. Science. 2009;325(5939):435–9. doi: 10.1126/science.1173288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West JD, Flockhart JH, Peters J, Ball ST. Death of mouse embryos that lack a functional gene for glucose phosphate isomerase. Genet Res. 1990;56(2–3):223–36. doi: 10.1017/s0016672300035321. [DOI] [PubMed] [Google Scholar]
- Yoshida Y, Takahashi K, Okita K, Ichisaka T, Yamanaka S. Hypoxia enhances the generation of induced pluripotent stem cells. Cell Stem Cell. 2009;5(3):237–41. doi: 10.1016/j.stem.2009.08.001. [DOI] [PubMed] [Google Scholar]
- Zheng P, Vassena R, Latham KE. Effects of in vitro oocyte maturation and embryo culture on the expression of glucose transporters, glucose metabolism and insulin signaling genes in rhesus monkey oocytes and preimplantation embryos. Mol Hum Reprod. 2007;13(6):361–71. doi: 10.1093/molehr/gam014. [DOI] [PubMed] [Google Scholar]
- Zhu S, Li W, Zhou H, Wei W, Ambasudhan R, Lin T, Kim J, Zhang K, Ding S. Reprogramming of human primary somatic cells by OCT4 and chemical compounds. Cell Stem Cell. 2010;7(6):651–5. doi: 10.1016/j.stem.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]