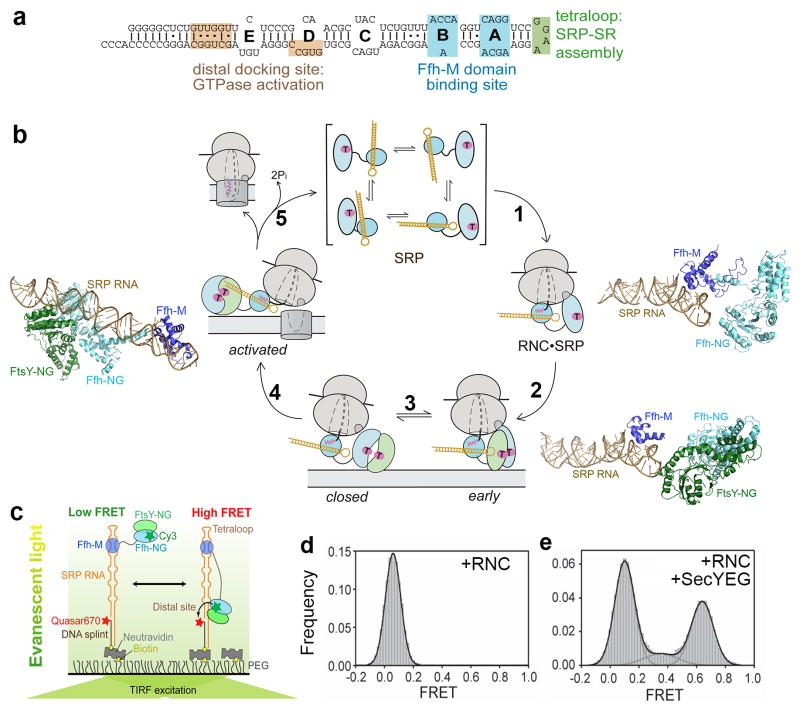
Figure 7.
SRP RNA-mediated global rearrangement of SRP couples cargo loading and unloading events to the GTPase cycle during protein targeting. (a) Secondary structure of the Escherichia coli 4.5S SRP RNA. The binding sites for the Ffh-M domain (blue) and for FtsY during the GTPase assembly (green) and activation (tan) steps are denoted. (b) Global rearrangement of SRP mediated by the SRP RNA during the protein targeting cycle. Top, free SRP exist in a variety of ‘latent’ conformations not conducive to the recruitment of FtsY. Right, binding of RNC induces a more active conformation of SRP (step 1), in which the SRP RNA tetraloop is properly positioned to interact with the G-domain of SR and hence form a stabilized early targeting complex (step 2). Molecular models derived from cyro-EM reconstructions are shown for the RNC•SRP (right panel) and RNC•SRP•SR early complex (lower right panel); the ribosome was not shown for clarity. Bottom, the GTPases detach from the SRP RNA tetraloop upon formation of the closed complex (step 3). Left, the GTPase complex relocalizes to the distal end of the SRP RNA (step 4), a conformation (left panel; PDB 2XXA) conducive to GTPase activation and cargo unloading (Step 5). All structures are aligned with respect to the SRP RNA. Color codings are the same as in Figure 3. The steps are numbered to be consistent with Figure 6a. (c) The smFRET setup to monitor the dynamic movements of the SRP-FtsY GTPase complex on the SRP RNA. FtsY Cys345 is labeled with Cy3, and the 5′end of the DNA splint is labeled with Quasar 670. (d–e) RNC and SecYEG regulate GTPase movement on the SRP RNA, as shown by the smFRET histograms of the SRP-FtsY complex bound to RNCFtsQ in the absence (d) and presence (e) of the SecYEG translocon. Adapted from reference (128).