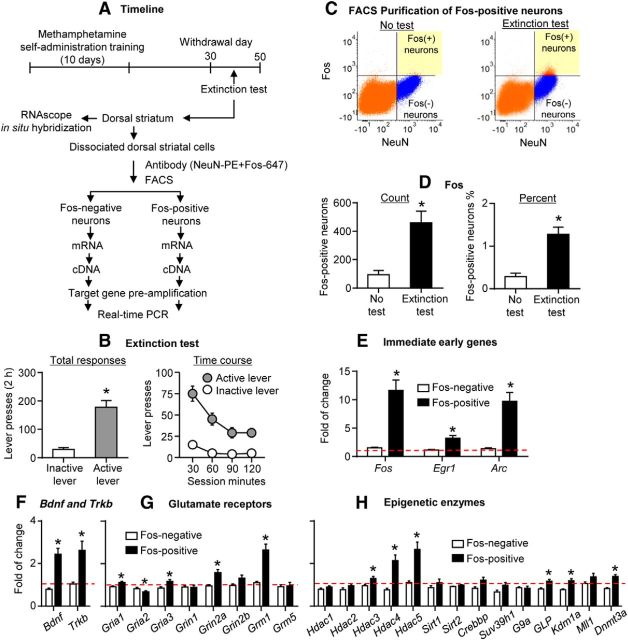
Figure 3.
Robust changes in mRNA expression of candidate genes in DS cue-activated Fos-positive neurons after prolonged withdrawal (Experiment 2, FACS). A, Timeline of the experiment. B, Extinction tests performed between withdrawal days 30 and 50. Data are the mean ± SEM of responses on the previously active lever and on the inactive lever during the 2 h extinction test. During testing, lever presses led to contingent presentations of the tone/light cue paired previously with methamphetamine infusions during training. *p < 0.001 (different from inactive lever); n = 15. C, An example of FACS sorting of NeuN-positive (x-axis) and Fos-positive or Fos-negative (y-axis) cells (Fos-positive and Fos-negative neurons) in DS. Left and right, Sorting of Fos-positive and Fos-negative neurons, respectively. Fos-positive and Fos-negative neurons are located in the top right and bottom right quadrants, respectively. D, FACS-sorted Fos-positive neurons. Left, Total number. Right, Percentage of total NeuN-positive neurons. *p < 0.01 (different from the No test group); n = 10–15 per group. E–H, Gene expression in FACS-sorted Fos-positive neurons in the Extinction test group. The immediate early genes (Fos, Egr1, Arc), Bdnf and Trkb, Gria 1, Gria3, Grin2a, Grm1, Hdac3, Hadc4, Hdac5, GLP, Kdm1a, and Dnmt3a increased. Gria2 decreased. Data are presented as folds of mean values in Fos-negative neurons from the No test group (red dashed line). *p < 0.005 (different from Fos-negative neurons). Error bars indicate SEM.