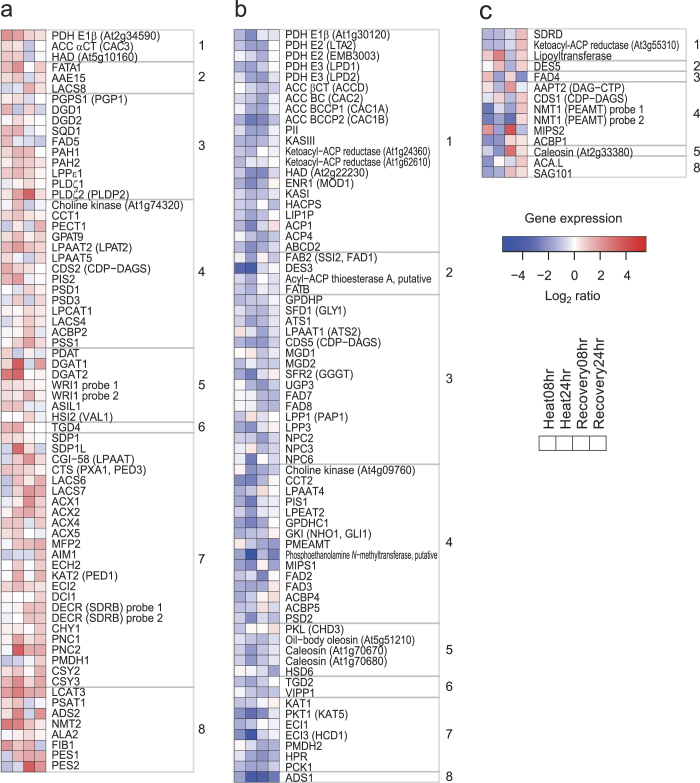
Figure 3. The expression of genes annotated to be involved in glycerolipid metabolism in Arabidopsis leaves under heat stress and during recovery.
The expression of 233 genes (228 probe sets) selected from the literature was analysed by microarray. Four sets of fold-changes were calculated as Heat08 hr/Control08 hr, Heat24 hr/Control24 hr, Recovery08 hr/Control08 hr and Recovery24 hr/Control24 hr and are shown as a heat map from left-to-right. (a) Probe sets increased >1.5-fold under at least one condition and not decreased, (b) probe sets decreased <0.5-fold under at least one condition and not increased, (c) probe sets both increased >1.5-fold under at least one condition and decreased <0.5-fold under at least one condition. Genes were selected from the literature, and classified into 8 groups of metabolic pathways: (1) plastidial fatty acid synthesis; (2) fatty acid elongation, desaturation and export from plastid; (3) plastidial glycerolipid, galactolipid and sulfolipid synthesis; (4) eukaryotic phospholipid synthesis; (5) TAG synthesis; (6) lipid trafficking; (7) TAG degradation, β-oxidation, the TCA/glyoxylate cycles; (8) miscellaneous lipid-related genes, and phytyl ester synthases. Genes that changed significantly (t-test with Benjamini-Hochberg FDR, p < 0.05, N = 3, 6, or 9) among the selected genes are shown. Further information is available in Supplementary Table S2.