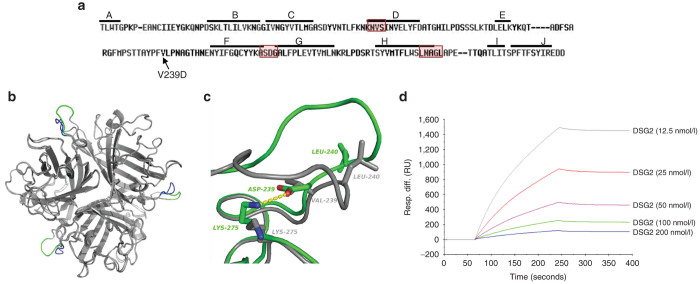
Figure 1.
Structural and functional studies with JO-4. (a) HAdV3 fiber knob amino acid sequence with β sheets A to J indicated by a line. JO-4 contains HAdV3 fiber knobs with a valine to aspartic acid substitution in position 239 of the HAdV3 fiber knob. Regions that have previously been found to be involved in DSG2 binding are boxed.22 (b) 3D-structure superimposition of the trimeric (wild type) HAdV3 fiber knob and the V239D mutated fiber knob. The common structure appears in grey, the EF loops of wild-type and mutant HAdV3 knob monomers are labeled in blue and green, respectively. (c) Shown is the interactions between D239 and K275 in the mutant and the new position of the L240 residue. Residue labels are colored green for the mutant and gray for wt. H-bonds between D239 and K275 are shown in dashes. (d) Affinity of JO-4 to DSG2 measured by surface plasmon resonance. JO-4 was immobilized on sensorchips, and background was automatically subtracted from the control flow cell. DSG2 were injected for 3 minutes at a concentration range from 12.5 to 200 nmol/l and kinetics and affinity parameters were evaluated using the BIAeval software (ka = 3.3 × 105 M−1s−1; kd = 4.7 × 10−4 s−1; KD =1.4 nmol/l).