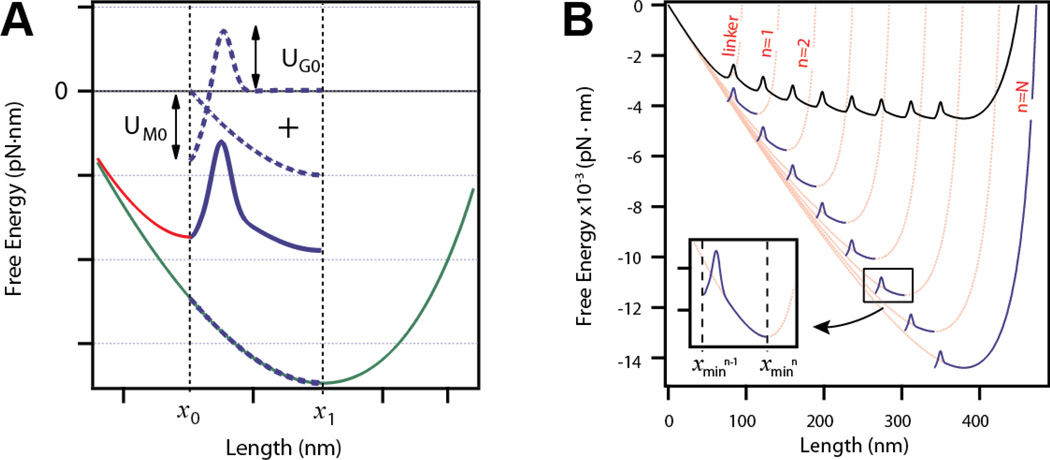
Figure 1. Method of the construction of the elastic free energy of a tandem modular protein.
A) Construction of the free energy for a single domain unfolding and extending. The free energy of unfolding and extending a domain is constructed from three elements (dotted lines): a Morse well of UM0 depth, a Gaussian barrier of UG0 height, and a WLC potential of contour length ΔLc (green line) from which only the section between x0 and x1 is considered. The red curve represents the initial extension to x0 calculated from the WLC with a contour length L0. B) Expansion of the free energy model to a polyprotein with N number of domains. The pink lines correspond to WLC curves calculated for an octamer polyprotein at a force F = 20 pN, and for contour length n ·ΔLc, where n is the number of unfolded domains, ΔLc = 19 nm, and p = 0.4 nm. At each contour length the WLC has a minimum that serves as the obligatory starting point for the next segment. Segments (thick blue lines) are constructed by adding a Morse well and a Gaussian barrier to the entropic elasticity as described in the text and in Figure 1A within the range xn−1 to xn (insert). The WLC for the linker is also shown in the plot (L0 =42 nm and pf =10 nm, extended at F = 20 pN). The final free energy profile is constructed by concatenating all segments at their boundaries (thick black line).