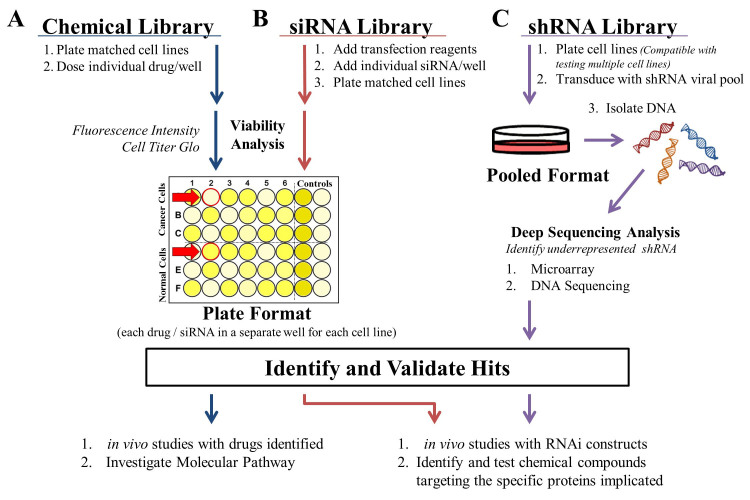
Figure 3.
Approaches to investigating synthetic lethal interactions. A) Chemical library screens are conducted using matched cell lines in a plate format wherein each drug is dosed separately to both lines. Fluorescence intensity from GFP or EYFP labeled cell lines can be used as a surrogate for cell count to make the assay high-throughput. B) siRNA library screens are also conducted in a plate format. Libraries are often custom ordered with individual siRNAs or siRNA pools and transfection reagent contained in each well. The matched cell lines can then be plated and following incubation analyzed in parallel using a high-throughput method like Cell Titer-Glo. Hits from screens performed using the plate format (A and B) are identified by comparing the normalized values of each well between the two matched cell lines. A hit, indicated by the red arrows, kills cancer cells while sparing the matched cell line (simulated normal tissue). C) The shRNA library approach is most often performed by transducing matched cell lines with a shRNA viral pool, although plated approaches can be conducted similar to the siRNA library approach. The pooled approach is more conducive for studying multiple cell lines and relies on deep sequencing analysis to identify hits. A hit is identified by identifying shRNAs that are lost after library transduction in cancer cell lines and present in matched (normal) cell lines, since synthetic lethal interactions will result in the cells death. Once hits have been identified and validated in each screen, the approaches A, B, C split. With a chemical library screen (A) in vivo studies can be readily initiated while the investigation into the molecular pathway is conducted. For siRNA and shRNA screens (B, C), in vivo studies can also be initiated using RNAi constructs while small molecules are developed.