Fig. 1. In vitro evolution of Notch1-DLL4 interactions with yeast surface display.
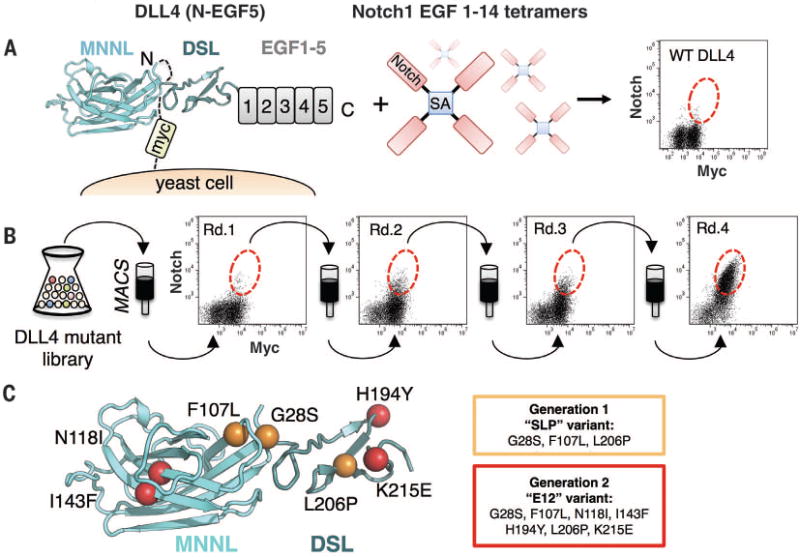
(A) Schematic representation of the yeast-displayed DLL4(N-EGF5)construct used for mutant library generation, stained with Notch1(1–14) tetramers. The flow cytometry dot plot shows double staining of DLL4 (N-EGF5) with 50 nM Notch1 tetramer and antibody to c-Myc. (B) Four rounds of selection were performed on magnetic-activated cell sorting (MACS) columns using a mutant library of DLL4(N-EGF5). Yeast isolated from selection rounds 1 to 4 (Rd.1 to Rd.4) were each double stained with 50 nM Notch1(1-14) tetramer and antibody to c-Myc. Dot plots for each round are displayed, with enriched populations circled with a red dotted line. (C) Affinity-enhancing mutants isolated from generation 1 and generation 2 libraries are mapped onto the surface of MNNL/DSL domains from the DLL4 crystal structure. The first generation SLP mutations are depicted as orange spheres. The second generation E12 variant includes the SLP mutations, plus four additional mutations (colored red).
