Figure 5.
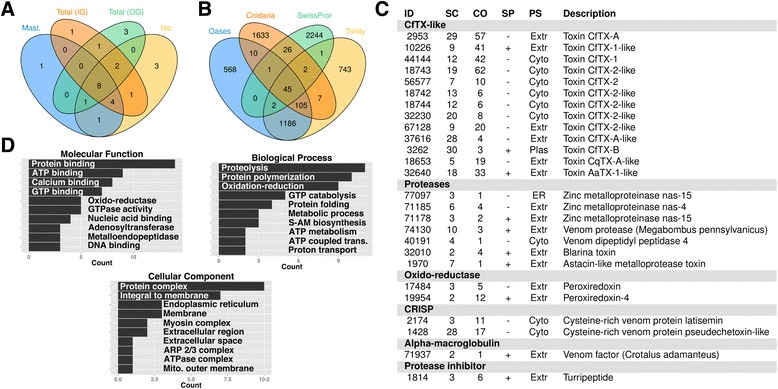
MS/MS analysis of C. fleckeri venom. A. Venn diagram showing the numbers of potential toxin proteins identified in each MS/MS experiment. Abbreviations used, Mast. — nematocyst sample containing predominantly mastigophores; Iso. — nematocyst sample containing predominantly isorhizas and trirhopaloids; Total (IG) — total nematocyst sample fractionated using SDS-PAGE; and Total (OG) — total nematocyst sample fractionated using peptide OFFGEL electrophoresis; B. Venn diagram showing overlap in significant peptide identifications in three additional databases searches using MS/MS data. The databases depicted are 1.) Oases — predicted protein dataset from Oases assembly; 2.) Trinity — predicted dataset Trinity assembly; 3.) Cnidaria — all cnidarian proteins from the GenBank non-redundant protein database; and 4.) SwissProt — the Uniprot SwissProt database; C. Proteins identified in the venom using MS/MS that had been previously identified as potential toxins during the analysis of the transcriptome; D. GO terms associated with proteins identified in the venom of C. fleckeri using MS/MS.
