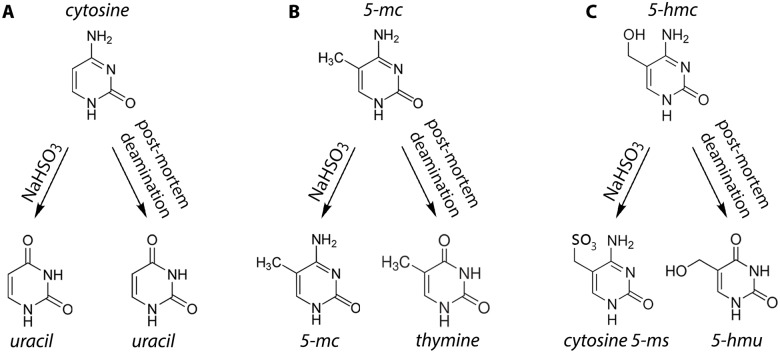
Fig 1. Detecting methylation of cytosine residues and their deamination products.
5-mc: 5-methylcytosine; 5-hmc: 5-hydroxymethylcytosine; 5-ms: 5-methylenesulfonate; 5-hmu: 5-hydroxymethyluracil; NaHSO3: Sodium bisulfite. (A) Unmethylated cytosines are converted to uracil at high efficiency by bisulfite conversion and at low efficiency by post-mortem deamination. After conversion, no methylation is detected by either bisulfite sequencing or misincorporation analysis. (B) Methylated cytosines are unaffected by bisulfite conversion, while post-mortem deamination converts methylated cytosines to thymines. Methylation is detected by the presence of undamaged cytosines in bisulfite sequencing, and by the presence of thymines at damaged positions in misincorporation analysis. (C) Hydroxymethylated cytosines are converted to cytosine 5-methelensulfonate by bisulfite conversion, and 5-hydroxymethyluracil by post-mortem deamination. Methylated cytosines are detected at undamaged positions by bisulfite sequencing, but cannot be discriminated from non-hydroxylated methylcytosines using this method. It is currently unclear whether misincorporation analysis will be able to detect methylation in the form of 5-hydroxymethyluracil, but the UDG-endoVIII approach may be able to do so.