Fig. 5.
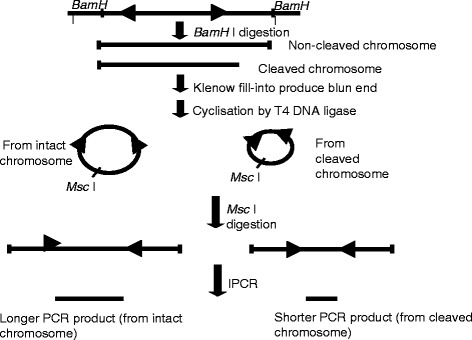
Flow chart showing DNA modification and IPCR. The arrow heads indicate the forward and reverse primers that were designed in opposite direction. BamH I digestion yielded a mixture of intact chromosome and cleaved chromosome. Klenow fill-in produced blunt ended chromosome fragments which were then cyclilsed by T4 DNA ligase. The intact chromosome will become a large circle while the cleaved chromosome will become a smaller circle. Upon cyclisation, the primers are now in correct orientation for amplification. Msc I digestion cleaved both circles outside the amplification region, thus merely linearise the molecule. Amplification from intact MLL gene will produce longer PCR products while amplification from cleaved MLL gene will yield shorter PCR products
