Figure 5.
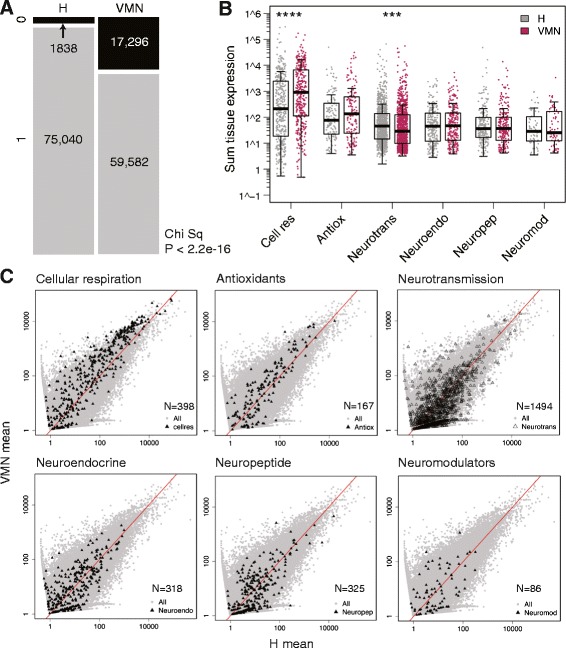
Tissue comparison of functional category expression. A) Raw counts from the brain transcriptome were dichotomized into expressed (1, gray) or not expressed (0, black) based on the summed tissue values. Compared to VMN, H has significantly more expressed genes. Chi sq: Chi squared test. B) Normalized abundances for each unique transcript expressed were summed from VMN or H sample groups, box-cox transformed, and subjected to one-way ANOVA for tissue comparison, with the Bonferroni corrected alpha level set at 0.008. Cellular respiration transcripts had significantly higher expression in the VMN while neurotransmission transcripts were higher in H. ****P < 0.0001, ***P = 0.0005. C) Mean VMN vs. H expression levels (fastlo-normalized) of transcripts within each of the six gene categories (black triangles) are plotted on top of all transcripts (gray dots). N: number of transcripts in each category. Cell res: cellular respiration; Antiox: antioxidants; Neurotrans: neurotransmission; Neuroendo: neuroendocrine; Neuropep: neuropeptide; Neuromod: neuromodulators.
