Abstract
Background
X-ray Computed Tomography (CT) imaging of the brain is commonly used in diagnostic settings. Although CT scans are primarily used in clinical practice, they are increasingly used in research. A fundamental processing step in brain imaging research is brain extraction – the process of separating the brain tissue from all other tissues. Methods for brain extraction have either been 1) validated but not fully automated, or 2) fully automated and informally proposed, but never formally validated.
Aim
To systematically analyze and validate the performance of FSL's brain extraction tool (BET) on head CT images of patients with intracranial hemorrhage. This was done by comparing the manual gold standard with the results of several versions of automatic brain extraction and by estimating the reliability of automated segmentation of longitudinal scans. The effects of the choice of BET parameters and data smoothing is studied and reported.
Methods
All images were thresholded using a 0 – 100 Hounsfield units (HU) range. In one variant of the pipeline, data were smoothed using a 3-dimensional Gaussian kernel (σ = 1mm3) and re-thresholded to 0 – 100 HU; in the other, data were not smoothed. BET was applied using 1 of 3 fractional intensity (FI) thresholds: 0.01, 0.1, or 0.35 and any holes in the brain mask were filled.
For validation against a manual segmentation, 36 images from patients with intracranial hemorrhage were selected from 19 different centers from the MISTIE (Minimally Invasive Surgery plus recombinant-tissue plasminogen activator for Intracerebral Evacuation) stroke trial. Intracranial masks of the brain were manually created by one expert CT reader. The resulting brain tissue masks were quantitatively compared to the manual segmentations using sensitivity, specificity, accuracy, and the Dice Similarity Index (DSI). Brain extraction performance across smoothing and FI thresholds was compared using the Wilcoxon signed-rank test. The intracranial volume (ICV) of each scan was estimated by multiplying the number of voxels in the brain mask by the dimensions of each voxel for that scan. From this, we calculated the ICV ratio comparing manual and automated segmentation: .
To estimate the performance in a large number of scans, brain masks were generated from the 6 BET pipelines for 1095 longitudinal scans from 129 patients. Failure rates were estimated from visual inspection. ICV of each scan was estimated and and an intraclass correlation (ICC) was estimated using a one-way ANOVA.
Results
Smoothing images improves brain extraction results using BET for all measures except specificity (all p < 0.01, uncorrected), irrespective of the FI threshold. Using an FI of 0.01 or 0.1 performed better than 0.35. Thus, all reported results refer only to smoothed data using an FI of 0.01 or 0.1. Using an FI of 0.01 had a higher median sensitivity (0.9901) than an FI of 0.1 (0.9884, median difference: 0.0014, p < 0.001), accuracy (0.9971 vs. 0.9971; median difference: 0.0001, p < 0.001), and DSI (0.9895 vs. 0.9894; median difference: 0.0004, p < 0.001) and lower specificity (0.9981 vs. 0.9982; median difference: −0.0001, p < 0.001). These measures are all very high indicating that a range of FI values may produce visually indistinguishable brain extractions. Using smoothed data and an FI of 0.01, the mean (SD) ICV ratio was 1.002 (0.008); the mean being close to 1 indicates the ICV estimates are similar for automated and manual segmentation.
In the 1095 longitudinal scans, this pipeline had a low failure rate (5.2%) and the ICC estimate was high (0.929, 95% CI: 0.91, 0.945) for successfully extracted brains.
Conclusion
BET performs well at brain extraction on thresholded, 1mm3 smoothed CT images with an FI of 0.01 or 0.1. Smoothing before applying BET is an important step not previously discussed in the literature. Analysis code is provided.
Keywords: CT, skull stripping, brain extraction, validation
1. Introduction
X-ray computed tomography (CT) scanning of the brain is widely available and is a commonly used diagnostic tool in clinical settings [1, 2, 3]. Though analysis of CT images is typically done by qualitative visual inspection, detailed quantification of information using neuroimaging tools is of interest. The reason for this interest is that qualitative inspection of CT scans provides limited quantifiable information that can be used in research. A fundamental processing step for producing quantifiable and reproducible information about the brain is to extract the brain from the CT image. This process is called brain extraction or skull stripping. This step is necessary because CT images contain non-brain human tissues and non-human elements (e.g. pillow, medical devices) that are not pertinent to brain research. We propose a validated automated solution to brain extraction in head CT scans using established neuroimaging software.
In magnetic resonance imaging (MRI), brain extraction has been extensively studied and investigated (see Wang et al. [4] for an overview of methods). While an extensive literature accompanied by software exist for brain MRI scans, the same is not true for brain CT scans. Smith [5] introduced and validated the Brain Extraction Tool (BET), a function of the FSL [6] neuroimaging software (v5.0.4), to automatically extract the brain from MRI scans. Here we propose to adapt BET and validate its brain extraction performance for CT scans.
BET adaptations for this purpose have been presented before in Solomon et al. [7]. Although the method is similar to that outlined below, using thresholding and then applying BET, the authors did not publish the specific details of the method nor any code to evaluate it. To replicate the method from Solomon et al. [7], Rorden et al. [8] thresholded voxels to be under 100 Hounsfield units, manually adjusted the image intensity to enhance the soft tissue in the brain, and then BET was applied with a fractional intensity of 0.35. Therefore, brain extraction methods from Rorden et al. [8] and Solomon et al. [7] strongly parallels the proposed method described below, but neither studies presented a formal validation against a set of manually segmented brain images.
Mandell et al. [9] has recently proposed a brain extraction method for CT scans and has done a validation against manual segmentation. This method was also performed on a set of brains with disease [10, 11]. This method is not fully automated, however. Mandell et al. [9] has formally validated a brain extraction method against manually segmented images, but the method requires user interaction.
Thus, the goals of our study are to propose an automated method that has been formally validated against a set of manually segmented images and estimate brain extraction performance of this method in a large number of CT scans.
2. Methods
2.1. Participants and CT data
We used CT images from patients enrolled in the MISTIE (Minimally Invasive Surgery plus recombinant-tissue plasminogen activator for Intracerebral Evacuation) and ICES (Intraoperative CT-Guided Endoscopic Surgery) stroke trials [12]. Inclusion criteria into the study included: 18 to 80 years of age, spontaneous supratentorial intracerebral hemorrhage above 20 milliliters (mL) in size (for full criteria, see Mould et al. [13]). The population analyzed here had a mean (SD) age was 60.6 (11.6) years, was 66.9% male, and was 55.6% Caucasian, 30.1% African American, 9.8% Hispanic, and 4.5% Asian or Pacific islander. CT data were collected as part of the Johns Hopkins Medicine IRB-approved MISTIE research studies with written consent from participants.
2.2. Imaging Data
2.2.1. Validation of Automated Head Segmentation
For the validation of automated segmentation against gold standard manual segmentation, we analyzed 36 scans, corresponding to 36 unique patients. The study protocol was executed with minor, but important, differences across the 19 sites. Scans were acquired using Siemens (N = 14), GE (N = 11), Philips (N = 10), and Toshiba (N = 1) scanners. Gantry tilt was observed in 21 scans. Slice thickness of the image varied within the scan for 7 scans. For example, a scan may have 10 millimeter (mm) slices at the top and bottom of the brain and 5mm slices in the middle of the brain. Therefore, the scans analyzed had different voxel (volume element) dimensions and image resolution prior to registration to the template. These conditions represent how scans are presented for evaluation in many diagnostic cases.
2.3. Manual and Automated Brain Extraction
Brain tissue was manually segmented as a binary mask from DICOM (Digital Imaging and Communications in Medicine) images using the OsiriX imaging software (OsiriX v.4.1, Pixmeo; Geneva, Switzerland) by one expert reader (reader 1: NU). CT brain images and the binary brain tissue mask obtained using manual segmentation were exported from OsiriX to DICOM format.
2.4. Image Processing
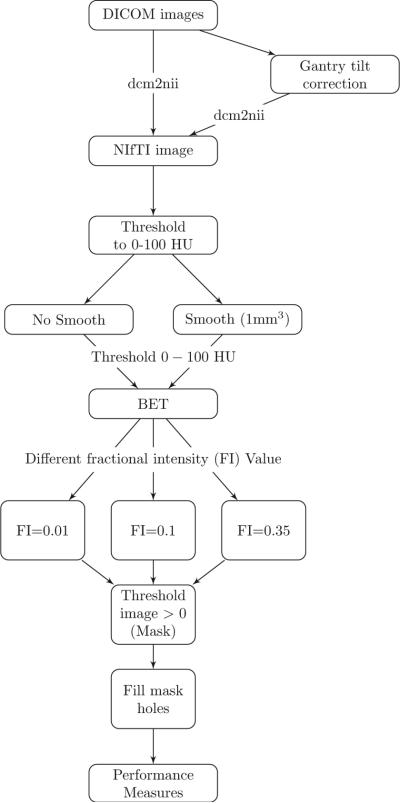
The image processing pipeline is provided in Figure 1. Images with gantry tilt were corrected using a customized MATLAB (The Mathworks, Natick, Massachusetts, USA) user-written script (http://bit.ly/1ltIM8c). Although gantry tilt correction is not inherently necessary for brain extraction, it is required for rigid co-registration of scans within a patient, which is a common processing step in longitudinal analysis of images post brain extraction.
Figure 1. Processing Pipeline.
Images in DICOM (Digital Imaging and Communications in Medicine) format were gantry tilt corrected if necessary and converted to NIfTI (Neuroimaging Informatics Technology Initiative) format using dcm2nii. After NIfTI conversion, the data is thresholded to tissue ranges of 0 – 100 Hounsfield units (HU). In one variant of the pipeline, the data was smoothed using a 3-dimensional Gaussian kernel (σ = 1mm3) and re-thresholded to 0 – 100 HU; in the other, the data was not smoothed. BET was applied to the image using 3 different fractional intensity (FI) values: 0.01, 0.1, and 0.35. The resultant image was masked to values greater than 0 HU and FSL was used to fill in any holes. These filled masks were used in comparison to the manually segmented image.
Images were converted to the Neuroimaging Informatics Technology Initiative (NIfTI) data format using dcm2nii (2009 version, provided with MRIcro [14]). Images were constrained to values between −1024 and 3071 HU to remove potential image rescaling errors and artifacts. No interpolation was done for images with a variable slice thickness. Thickness was determined from the first converted slice and the NIfTI format assumes homogeneous thickness throughout the image. This loss of information, if not properly accounted for, affects volume estimation, which relies on accurate pixel dimensions in millimeters. Variable slice thickness should have no affect on the other estimates of performance described below as they are calculated at a voxel level and do not rely on pixel resolution. Although the NIfTI images store the data with only one pixel dimension for the height of the voxel, we use the ImagePositionPatient DICOM field to determine the accurate height of each voxel to calculate an accurate volume.
Each image was thresholded using the brain tissue range (0 – 100 HU); voxels outside this range were set to 0 HU. In one variant of the pipeline, data were smoothed using a 3-dimensional (3D) Gaussian kernel (σ = 1mm3) and re-thresholded to 0−100 HU; in the other, data were not smoothed. BET was then applied, varying the fractional intensity (FI) parameter to determine its influence on performance: we used values of 0.35 (as used in Rorden et al. [8]), 0.1, and 0.01.
The FI parameter varies between 0 and 1 and determines the location of the edge of the segmented brain image; smaller values correspond to larger brain masks. Smith [5] describes that the FI parameter determines a local threshold tl by the following equation:
where Imax is a local maximum intensity along a line from an outer surface vertex pointing inward to the image center and t2 is the 2nd percentile of the image distribution. As a result of thresholding in our pipeline, t2 equals 0 HU and Imax must lie between 0 and 100 HU. Therefore, after thresholding,
With an Imax of 100 HU, using an FI lower than 0.01 results in t1 less than 1 HU, but greater than 0 HU. As CT data is stored as integers, no intensities lie between 0 and 1, so a t1 between 0 or 1 HU should provide similar local thresholds. Therefore, we chose 0.01 as a lower limit for testing FI.
After BET was applied, we created a brain mask taking values > 0 HU and filled the holes in the mask (using fslmaths -fillh).
2.5. Measuring and Testing Brain Extraction Performance
We compared the masks obtained using the various choices of parameters to the manually segmented images. Four common measurements of performance were calculated for each image: sensitivity, specificity, accuracy, and the Dice Similarity Index (DSI) [15]. For each measure, higher values indicate better agreement with the manual segmentation. See Inline Supplementary Methods 1 for the calculation of each measure.
We calculated the paired difference of each measure using different pipelines (e.g. 0.01 vs. 0.1, smoothed data). We tested whether these differences were statistically different from zero using the Wilcoxon signed-rank test.
From each scan, we also calculated the intracranial volume (ICV), defined as all voxels inside the skull, by multiplying the number of voxels in the resulting mask by the dimensions of each voxel. We calculated the ICV ratio comparing manual and automated segmentation: . A ratio of 1 indicates the same volume; greater than 1 indicates over-estimation of ICV; less than 1 indicates underestimation of ICV. As adjustment for ICV has been shown to reduce inter-subject variation in volumetric studies [16], we wish to estimate ICV accurately.
2.6. Consistency of Manual Brain Extraction
As manual segmentation can have intra-reader variability, another reader (reader 2: AM) manually segmented brain tissue on the 36 scans. We additionally estimated all four performance measurements, using the manual segmentation from reader 2 as the gold standard. We also estimated the ICV from the segmentation from reader 2. We calculated the ICV ratio and the correlation of ICV estimates across readers.
2.7. Failure Rate and Intraclass Correlation Estimate
Although comparison of automated methods to a manual gold standard is ideal, manual segmentation requires a significant amount of time. Therefore, for a large number of scans, this procedure is impractical. As multiple CT scans are obtained from patients in the MISTIE trial, we can estimate the reliability of our proposed brain extraction pipelines without manual segmentation by comparing intracranial volumes of the same patient on subsequent scans. Moreover, we can estimate failure rate of each pipeline.
For these tasks, we collected 1160 scans. Of these scans, we excluded 27 scans due to craniotomy and 38 due to the gantry tilt correction forcing areas of the brain outside the field of view. We executed the previous brain extraction pipelines on the remaining 1095 scans. Of these scans, we visually assessed the quality of brain extraction: any scan excluding a significant portion of the brain or having holes due to mask self-intersection were classified as a failure. These scans represent 129 patients from 26 sites, with a mean (SD) of 8.5 (2.8) scans per patient. Scans were acquired using Siemens (N = 492), GE (N = 298), Philips (N = 207), Toshiba (N = 66), Neurologica (N = 30), and Picker (N = 2) scanners. We estimated the failure rate for each processing pipeline and used a Fisher's exact test to test whether failure rates differed across scanners.
For each scan, we calculated the ICV. Using only the scans with successful brain extraction, we estimated the intraclass correlation (ICC) and its confidence interval (CI) of ICV using a one-way ANOVA, where a patients was treated as a group, for unbalanced repeated measures [17, 18, 19, 20] using the ICC package [21] in R (http://cran.r-project.org/).
3. Results
3.1. Manual and Automated Brain Extraction
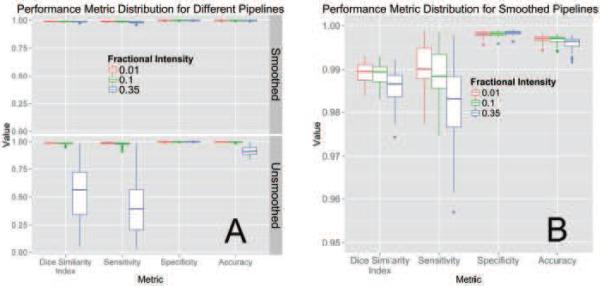
The following estimates use the manual segmentation from reader 1 as the gold standard. Figure 2A illustrates the performance of each variation of the BET pipeline in Figure 1. The pipelines using smoothing (top panel) perform better than the unsmoothed pipelines (bottom panel) on all measures except specificity (all p < 0.01, uncorrected for multiplicity). BET also performed poorly on some scans without smoothing.
Figure 2. Performance Metric Distribution for Different Pipelines.
Panel A displays the boxplots for performance measures when running the pipeline with a different fractional intensity (FI), using smoothed data (top) or unsmoothed data (bottom). Panel B presents the smoothed data only, rescaled to show discrimination between the different FI. Overall, FI of 0.01 and 0.1 perform better than 0.35 in all categories other than specificity. Using smoothed data improves performance in all performance metrics, markedly when an FI of 0.35 is used. Panel B demonstrates that using an FI of 0.01 on smoothed data has high performance on all measures.
Figure 2B displays the performance for brain extraction for the pipelines using smoothed images. Because the performance for all metrics was high when using smoothed images, it was necessary to change the y-axis from [0, 1] to [0.95, 1]. Using an FI of 0.01 or 0.1 performed better than 0.35; thus, we will focus and compare results for these values of FI only for the case when BET was applied to smoothed images. Using an FI of 0.01 had a higher median sensitivity (0.9901) than an FI of 0.1 (0.9884, median difference: 0.0014, p < 0.001), accuracy (0.9971 vs. 0.9971; median difference: 0.0001, p < 0.001), and DSI (0.9895 vs. 0.9894; median difference: 0.0004, p < 0.001) and lower specificity (0.9981 vs. 0.9982; median difference: −0.0001, p < 0.001). Overall, regardless of p-values, these measures are all very high, indicating that multiple choices of parameters work well for brain extraction after CT image processing. Moreover, a Bonferroni correction for multiple comparisons yields the same conclusions.
The mean (SD) ICV ratio was 1.002 (0.0079) using an FI of 0.01 and 1 (0.0081) using an FI of 0.1. Both mean ratios are close to 1 with a small variance, indicating the ICV estimates are similar for automated and manual segmentation.
The above results indicate that using smoothed data and an FI of 0.01 or 0.1 had high performance when compared to the manual segmentation of reader 1. The results were similar using the scan-wise union of the segmentation from reader 1 and reader 2. Using the manual segmentation from reader 2 or the scan-wise intersection of the segmentation from reader 1 and reader 2, the median values using and 0.1 had higher marginally performance than using 0.01 for DSI, accuracy, and specificity, but lower performance for sensitivity. See Inline Supplementary Figure 1 for the distribution of performance metrics for each segmentation.
Regardless of which manual segmentation was used, estimates of performance for each scan using smoothed data and an FI of 0.01 or 0.1 remained above 0.95. Thus, these pipelines perform well, yet one FI may not perform universally better than the other.
3.2. Consistency of Manual Brain Extraction
When comparing manual segmentations, we used the manual segmentation from reader 1 as the gold standard and the segmentation from reader 2 as the test segmentation similar to the automated segmentation above. The mean (SD) was 0.989 (0.0030) for DSI, 0.997 (0.0010) for accuracy, 0.982 (0.0060) for sensitivity, and 0.999 (0.0003) for specificity.
The estimated mean (SD) ratio of the ICV was 0.988 (0.0068). The correlation (95% confidence interval) of ICV was 0.998 (0.997, 0.999). See Inline Supplementary Figure 2 for the comparison of ICV estimates from reader 1 and reader 2.
Overall, we observe high agreement of segmentation between raters and the estimates of performance in automated segmentation to be similar to multiple reader segmentation. Differences between manual segmentation occurred on the boundary between bone and non-bone areas towards the surface of the cortex and inferior regions of the brain, where one may or may not classify areas as spinal cord and not part of the brain stem. Thus, the difference observed in the performance of FI of 0.01 compared to 0.1 when using a reader 2 as the gold standard are likely due to these areas.
3.3. Failure Rate and Intraclass Correlation Estimate
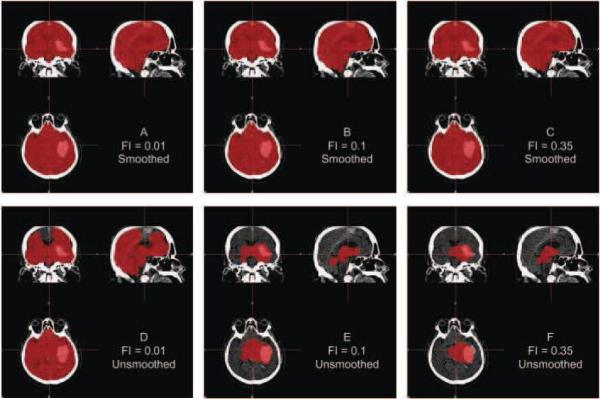
Although Figure 2 indicates that using FI of 0.01 or 0.1 provides adequate brain extraction results for the cases analyzed, they perform relatively well regardless whether or not the data are smoothed. Figure 3 displays an example where using unsmoothed data performs poorly for these FIs, demonstrating why smoothing may be necessary for some scans. This is a high-resolution scan, with voxel size 0.49mm×0.49mm×1, which may result in more noise in the image that may affect the performance of BET. Moreover, in Table 1, the estimated failure rates were lower using the smoothed data compared to the unsmoothed data. We observe the lowest failure rate in the pipelines using smoothed data and an FI of 0.01 or 0.1. Though this represents a large number of scans, failure rates may also be affected by patient-level characteristics, including the center where the patient was scanned.
Figure 3. Example Case where Smoothing before BET is Required.
For one subject, the CT image is displayed with the brain extracted mask in red after running all pipelines. Panels A, B, and C represent applying BET using FI of 0.01, 0.1, and 0.35, respectively, to smoothed data. Panels D, E, and F correspond to applying BET using FI 0.01, 0.1, and 0.35 on unsmoothed data. Smoothing images improves brain extraction with BET.
Table 1.
Failure Rates for each Processing Pipeline of Brain Extraction of the 1095 Scans Analyzed.
| Failure Scans: N (%) | ||
|---|---|---|
| Fractional Intensity | Unsmoothed | Smoothed |
| 0.01 | 161 (14.7%) | 57 (5.2%) |
| 0.1 | 192 (17.5%) | 80 (7.3%) |
| 0.35 | 1068 (97.5%) | 154 (14.1%) |
As multiple scanners were used, we wanted to determine if the failure rate was different across scanners. In Table 2, we present the failure rate for each scanner, using smoothed data and an FI of 0.01. The failure rates for all scanner types other than Neurologica were below 6%. Although we see a failure rate above 16% in the Neurologica scanners, only 30 scans were acquired using this type of scanner. Moreover, a Fisher's exact test did not find a difference in the failure rates across scanner type (p = 0.110).
Table 2.
Failure Rates for Different Scanner Types using Smoothed Data and an FI of 0.01 Processing Pipeline of Brain Extraction of the 1095 Scans Analyzed.
| Scanner Type | Failure Rate: Fail/N (%) |
|---|---|
| Siemens | 28/492 (5.7%) |
| GE | 15/298 (5.0%) |
| Philips | 7/207 (3.4%) |
| Toshiba | 2/66 (3.0%) |
| Neurologica | 5/30 (16.7%) |
| Picker | 0/2 (0.0%) |
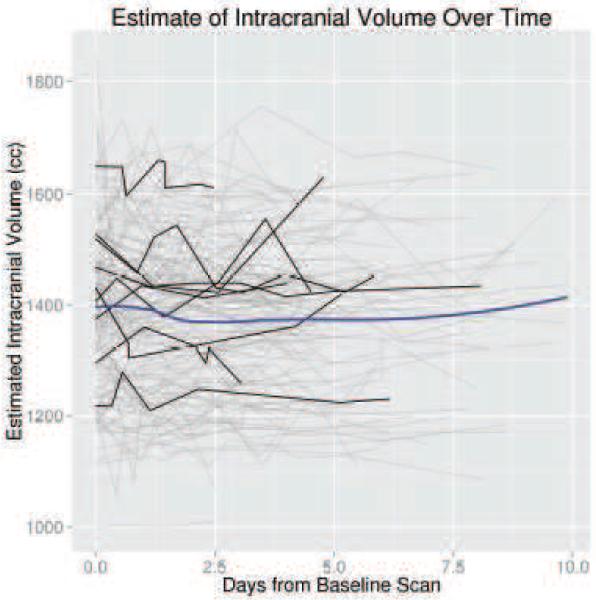
The ICC estimate was high using the successfully brain extracted scans from the smoothed data with an FI of 0.01 (ICC: 0.929, 95% CI: 0.91, 0.945) and 0.1 (ICC: 0.928, 95% CI: 0.909, 0.944). In Figure 4, we illustrate the ICV estimates, using an FI of 0.01 and smoothed data, for successful brain extraction in scans 10 or fewer days post baseline scan (gray lines). The black lines represent ICV estimates over time for 10 randomly selected patients. The blue line is a local regression (LOESS) [22] line, which represents an estimate of the average ICV over time. This LOESS line is relatively flat, indicating that the ICV estimate averaged over patients is stable. We also observe that although within-patient variability exists for ICV estimates, the variability across patients is greater.
Figure 4. Intracranial Volume (ICV) Estimates for Scans Less than 10 Days Post-Baseline.
These estimates are from the brain extraction pipeline using and FI of 0.01 and smoothed data. Each separate line represents an individual patient. The black lines represent ICV estimates over time for 10 randomly selected patients. The blue line is a local regression (LOESS) scatterplot smoother.
4. Discussion
Quantitative procedures based on data contained in head CT images require brain-only images for analysis. We have introduced the first validated, fully automated brain extraction pipeline for head CT images based on widely used, existing software. Validation was done using gold-standard manual segmentations of brain tissue and multiple measurements of intracranial volume per patient. A novel finding is that smoothing the data, as opposed to other studies which have smoothed at other points in the process [9], using a conservative smoother (1mm3 3D Gaussian kernel) and using an FI of 0.01 or 0.1 provides good brain extraction for the sample studied. These choices make a large difference in the performance of the algorithms and have not been previously reported in the literature.
Although the sample size was relatively small for the gold standard validation, the CT images used are from different people, different centers, and different scanners. We have also shown that failure rates are low (5%) using smoothed data and an FI of 0.01 in a large number of scans. We are using a population of patients with intracranial hemorrhage and the accuracy of BET may be dependent on factors such as hematoma size, which may change the distribution of Hounsfield units or compress brain structures. We observed good performance of BET in these patients using the parameters described, which may indicate even better performance for individuals with no observed pathology. BET also performs well in scans from follow-up scans with no hemorrhage included in the longitudinal ICV estimates; these scans should be more similar to scans from patients without hemorrhage.
We did not, however, rigorously test this pipeline against a set of different levels of noise (as was done using MRI in [9]), convolution filters, scanning artifacts, or scanning parameters. As noted in Mandell et al. [9], there is no standard ground truth for CT scans. However, as these scans are those presented for evaluation in many diagnostic cases, we believe these do represent clinically relevant scans that would be analyzed. As the failure rates are low, we believe this method may be adapted for different settings of acquisition for CT.
A percentage of scans in this study did not have successful brain extraction, however. Additional steps may be done to decrease the failure rate of the proposed method: 1) using registration to a CT template [8] to remove slices of the neck below the head can achieve better brain-center estimation used by BET; 2) performing BET using a higher smoothness constraint that may reduce potential holes in the brain mask caused by mask intersection; and 3) using a dilation followed by an erosion operation to fill any holes that are caused by thresholding but not filled by the hole filling operation above.
One additional concern is how general this method is to other populations as the data is from an older adult population, such as the pediatric population [10] analyzed. Gousias et al. [23] analyzed 33 2-year-old children who had been born prematurely and found that after preprocessing of co-registration and neck removal, and dilation, BET was adequate for brain extraction in MR images, and then brain labeling. Moreover, Shi et al. [24] demonstrated BET performed well in automated brain extraction for pediatric MR images of neonates ¡ 2 months (N = 90), 1-2 year infants (N = 141) and 5-18 year old children (N = 60). Although CT scans may have differences compared to MRI, we believe this method should be robust at least to children, but would like to validate our method on additional populations.
Overall, good performance using CT acquired under different scanners and different scanning parameters indicate that the approach described here will likely generalize in addition to the fact that CT scan data are expressed in standardized units (Hounsfield units). Moreover, the robust success of BET as a method is another indicator that our proposed method has a high likelihood of generalizability.
After creating an accurate brain mask, secondary image processing or estimation steps can be performed. These include intensity normalization, segmentation, and image registration. Moreover, ICV estimates can be used as potential factor for adjustment in analysis [16]. Additionally, extraction of structures within the brain, such the cerebrospinal fluid (CSF), which can be estimated from the method described in Volkau et al. [25], may have fewer errors if performed on the segmented image as the process is not computed over voxels outside of the brain. We believe that successful brain extraction is fundamental for calculating quantitative measures on the brain and performing necessary secondary operations required for analysis.
The research presented here is fully reproducible and we provide ready-to-use software for CT brain extraction. The R function designed to perform brain extraction is located at http://bit.ly/CTBET_RCODE and example bash script for command-line FSL can be downloaded here http://bit.ly/CTBET_BASH. As our software is publicly available and is based on open-source, free programs (FSL and R), the proposed method is readily available to all users.
Supplementary Material
Highlights.
- We present a fully automated brain extraction procedure for CT scans adapting the BET from FSL
- We investigated the effect smoothing the data before using BET; smoothing improves performance
- We varied the fractional intensity using 0.01, 0.1, and 0.35. 0.01 and 0.1 performed best
- We validated against a gold standard – accuracy and Dice overlap of automated segmentation was high
Acknowledgements
We thank the patients and families who volunteered for this study and Genentech Inc. for the donation of the study drug (Alteplase).
Sources of Funding
The project described was supported by the NIH grant RO1EB012547 from the National Institute of Biomedical Imaging And Bioengineering, T32AG000247 from the National Institute on Aging, R01NS046309, RO1NS060910, RO1NS085211, R01NS046309, U01NS080824 and U01NS062851 from the National Institute of Neurological Disorders and Stroke, and RO1MH095836 from the National Institute of Mental Health. Minimally Invasive Surgery and rt-PA in ICH Evacuation Phase II (MISTIE II) was supported by grants R01NS046309 and U01NS062851 awarded to Dr. Daniel Hanley from the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS). ICES was led by Co-Principal Investigator Dr. Paul Vespa at the University of California Los Angeles. Minimally Invasive Surgery and rt-PA in ICH Evacuation Phase III (MISTIE III) is supported by the grant U01 NS080824 awarded to Dr. Daniel Hanley from the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS). Clot Lysis: Evaluating Accelerated Resolution of Intraventricular Hemorrhage Phase III (CLEAR III) is supported by the grant U01 NS062851 awarded to Dr. Daniel Hanley from the National Institutes of Health (NIH)/National Institute of Neurological Disorders and Stroke (NINDS).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Sahni R, Weinberger J. Management of intracerebral hemorrhage. Vascular Health and Risk Management. 2007;3(5):701–709. ISSN 1176-6344. [PMC free article] [PubMed] [Google Scholar]
- 2.Chalela JA, Kidwell CS, Nentwich LM, Luby M, Butman JA, Demchuk AM, Hill MD, Patronas N, Latour L, Warach S. Magnetic resonance imaging and computed tomography in emergency assessment of patients with suspected acute stroke: a prospective comparison. The Lancet. 2007;369(9558):293–298. doi: 10.1016/S0140-6736(07)60151-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schellinger PD, Jansen O, Fiebach JB, Hacke W, Sartor K. A standardized MRI stroke protocol comparison with CT in hyperacute intracerebral hemorrhage. Stroke. 1999;30(4):765–768. doi: 10.1161/01.str.30.4.765. [DOI] [PubMed] [Google Scholar]
- 4.Wang Y, Nie J, Yap P-T, Li G, Shi F, Geng X, Guo L, Shen D, Initiative ADN. Knowledge-guided robust mri brain extraction for diverse large-scale neuroimaging studies on humans and non-human primates. PloS one. 2014;9(1):e77810. doi: 10.1371/journal.pone.0077810. others. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith SM. Fast robust automated brain extraction. Human Brain Mapping. 2002;17(3):143–155. doi: 10.1002/hbm.10062. ISSN 1097-0193, doi:10.1002/hbm.10062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jenkinson M, Beckmann CF, Behrens TEJ, Woolrich MW, Smith SM. FSL. NeuroImage. 2012;62(2):782–790. doi: 10.1016/j.neuroimage.2011.09.015. ISSN 1053-8119, doi:10.1016/j.neuroimage.2011.09.015. [DOI] [PubMed] [Google Scholar]
- 7.Solomon J, Raymont V, Braun A, Butman JA, Grafman J. User-friendly software for the analysis of brain lesions (ABLe) Computer Methods and Programs in Biomedicine. 2007;86(3):245–254. doi: 10.1016/j.cmpb.2007.02.006. ISSN 0169-2607, doi:10.1016/j.cmpb.2007.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rorden C, Bonilha L, Fridriksson J, Bender B, Karnath H-O. Age-specific CT and MRI templates for spatial normalization. NeuroImage. 2012;61(4):957–965. doi: 10.1016/j.neuroimage.2012.03.020. ISSN 1053-8119, doi:10.1016/j.neuroimage.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mandell JG, Langelaan JW, Webb AG, Schiff SJ. Volumetric brain analysis in neurosurgery: Part 1. Particle filter segmentation of brain and cerebrospinal fluid growth dynamics from MRI and CT images. Journal of Neurosurgery: Pediatrics. 2014:1–12. doi: 10.3171/2014.9.PEDS12426. [DOI] [PubMed] [Google Scholar]
- 10.Mandell JG, Kulkarni AV, Warf BC, Schiff SJ. Volumetric brain analysis in neurosurgery: Part 2. Brain and CSF volumes discriminate neurocognitive outcomes in hydrocephalus. Journal of Neurosurgery: Pediatrics. 2014:1–8. doi: 10.3171/2014.9.PEDS12427. [DOI] [PubMed] [Google Scholar]
- 11.Mandell JG, Hill KL, Nguyen DT, Moser KW, Harbaugh RE, McInerney J, Nsubuga BK, Mugamba JK, Johnson D, Warf BC, et al. Volumetric brain analysis in neurosurgery: Part 3. Volumetric CT analysis as a predictor of seizure outcome following temporal lobectomy. Journal of Neurosurgery: Pediatrics. 2014:1–11. doi: 10.3171/2014.9.PEDS12428. [DOI] [PubMed] [Google Scholar]
- 12.Morgan T, Zuccarello M, Narayan R, Keyl P, Lane K, Hanley D. Cerebral Hemorrhage. Springer; 2008. Preliminary findings of the minimally-invasive surgery plus rtPA for intracerebral hemorrhage evacuation (MISTIE) clinical trial; pp. 147–151. [DOI] [PubMed] [Google Scholar]
- 13.Mould WA, Carhuapoma JR, Muschelli J, Lane K, Morgan TC, McBee NA, Bistran-Hall AJ, Ullman NL, Vespa P, Martin NA, Awad I, Zuccarello M, Hanley DF. Minimally Invasive Surgery Plus Recombinant Tissue-type Plasminogen Activator for Intracerebral Hemorrhage Evacuation Decreases Perihematomal Edema. Stroke. 2013;44(3):627–634. doi: 10.1161/STROKEAHA.111.000411. ISSN 0039-2499, 1524-4628, doi:10.1161/STROKEAHA.111.000411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rorden C, Brett M. Stereotaxic Display of Brain Lesions. Behavioural Neurology. 2000;12(4):191–200. doi: 10.1155/2000/421719. ISSN 0953-4180, doi:10.1155/2000/421719. [DOI] [PubMed] [Google Scholar]
- 15.Dice LR. Measures of the amount of ecologic association between species. Ecology. 1945;26(3):297–302. [Google Scholar]
- 16.Whitwell JL, Crum WR, Watt HC, Fox NC. Normalization of cerebral volumes by use of intracranial volume: implications for longitudinal quantitative MR imaging. American Journal of Neuroradiology. 2001;22(8):1483–1489. [PMC free article] [PubMed] [Google Scholar]
- 17.Searle SR. Linear models. John Wiley & Sons; 2012. [Google Scholar]
- 18.Thomas JD, Hultquist RA. Interval estimation for the unbalanced case of the one-way random effects model. The Annals of Statistics. 1978:582–587. [Google Scholar]
- 19.Donner A. The use of correlation and regression in the analysis of family resemblance. American journal of epidemiology. 1979;110(3):335–342. doi: 10.1093/oxfordjournals.aje.a112819. [DOI] [PubMed] [Google Scholar]
- 20.Lessells CM, Boag PT. Unrepeatable repeatabilities: a common mistake. The Auk. 1987:116–121. [Google Scholar]
- 21.Wolak ME, Fairbairn DJ, Paulsen YR. Guidelines for estimating repeatability. Methods in Ecology and Evolution. 2012;3(1):129–137. ISSN 2041-210X, doi:10.1111/j.2041-210X.2011.00125.x. [Google Scholar]
- 22.Cleveland WS, Grosse E, Shyu WM. Local regression models. Statistical models in S. 1992:309–376. [Google Scholar]
- 23.Gousias IS, Rueckert D, Heckemann RA, Dyet LE, Boardman JP, Edwards AD, Hammers A. Automatic segmentation of brain MRIs of 2-year-olds into 83 regions of interest. Neuroimage. 2008;40(2):672–684. doi: 10.1016/j.neuroimage.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 24.Shi F, Wang L, Dai Y, Gilmore JH, Lin W, Shen D. LABEL: pediatric brain extraction using learning-based meta-algorithm. Neuroimage. 2012;62(3):1975–1986. doi: 10.1016/j.neuroimage.2012.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Volkau I, Puspitasari F, Nowinski WL. Ventricle boundary in CT: partial volume effect and local thresholds. Journal of Biomedical Imaging. 2010;2010:15. doi: 10.1155/2010/674582. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.