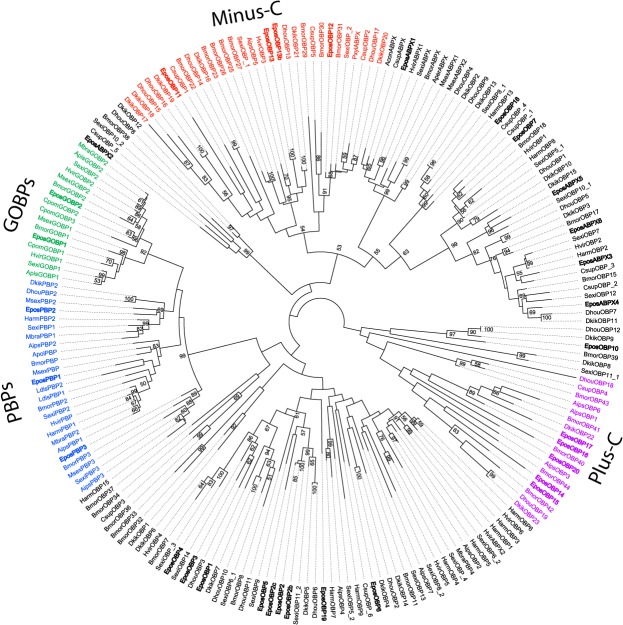
Fig 1. A maximum likelihood phylogenetic tree of odorant binding proteins (OBPs) from moths.
Includes sequences from Epiphyas postvittana (Epos), Bombyx mori (Bmor), Cydia pomonella (Cpom), Chilo suppressalis (Csup), Dendrolimus houi (Dhou), Dendrolimus kikuchii (Dkik), Helicoverpa armigera (Harm), Heliothis virescens (Hvir), Lymantria dispar (Ldis), Mamastra brassicae (Mbra), Manduca sexta (Msex), Plutella xylostella (Pxyl) and Spodoptera exigua (Sexi). OBPs identified from E. postvittana are in bold. Node support was assessed using bootstrap replicates of 1000 and values greater than 50% are shown. Sub-groups including the PBPs, GOBPs, minus-Cs and plus-Cs are marked.