Figure 3.
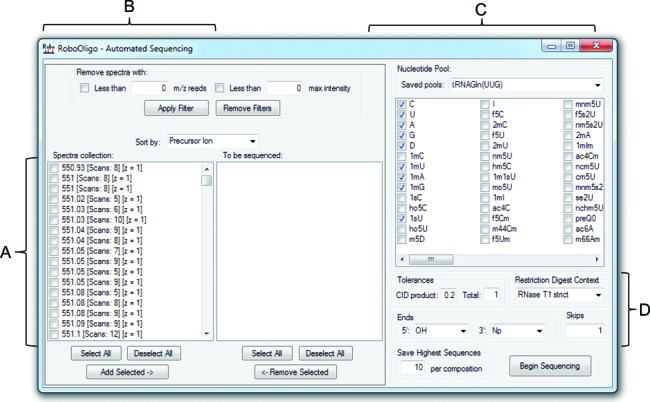
Automated de novo sequencing. (A) MS/MS data can be selected and moved to the ‘To be sequenced’ box to be included in the analysis. (B) MS/MS data with less than a user selected number of m/z reads and/or maximum ion abundance can be removed from the ‘Scan collection’ box. (C) Nucleotides to be included in the automated de novo sequencing attempt—oligomers of <8 nt can utilize pools of up to 20 nt, while longer oligomers return the best results with pools of <12 nt. New entries in the ‘Saved pools’ dropdown list can be added by editing the ‘NucleotidePools.txt’ file. (D) The CID m/z tolerance, total mass tolerance, RNase digestion context, 5′ and 3′ ends and number of allowed skips are all modifiable by the user.
