Figure 3.
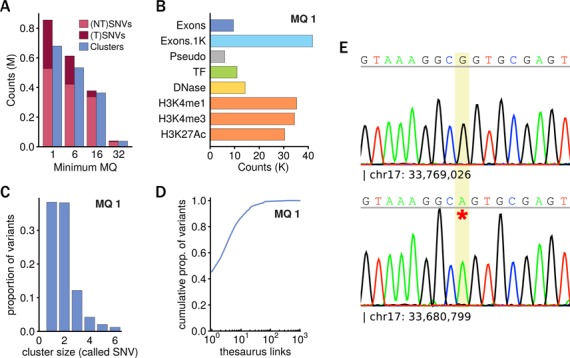
Thesaurus analysis of genome sequencing data from a haploid cell line. (A) Variants called with several mapping quality (MQ) thresholds were thesaurus-filtered and grouped into clusters. (T)SNVs and (NT)SNVs refer to sites that were or were not annotated with links to alternate sites, respectively. Variants that were not thesaurus-annotated and were common to all call sets were excluded from the plot (2.5M variants). (B) Many thesaurus-annotated variants called with lenient MQ threshold fall in candidate functional regions as defined by ENCODE (19). Exons: exonic regions defined by Gencode (V19); Exons.1K: exonic regions plus 1 kb flanking regions; Pseudo: exonic regions labeled by PseudoGene; TF: transcription factor binding sites in various cell types; DNase: DNase hypersensitivity clusters in various cell types; H3K4me1: histone methylation mark in seven cell types; H3K4me3: histone trimethylation mark in seven cell types; H3K27Ac: histone acetylation marks in seven cell types. (C) Cluster sizes formed by called exonic variants linked together by thesaurus annotation. (D) Number of alternate sites associated with called variants in exons. (E) Two Sanger traces produced by different specific primers. Top trace shows a wild-type sequence within gene SLFN13; bottom trace shows sequence within gene SLFN11 with a homozygous variant at the indicated position.
