Figure 4.
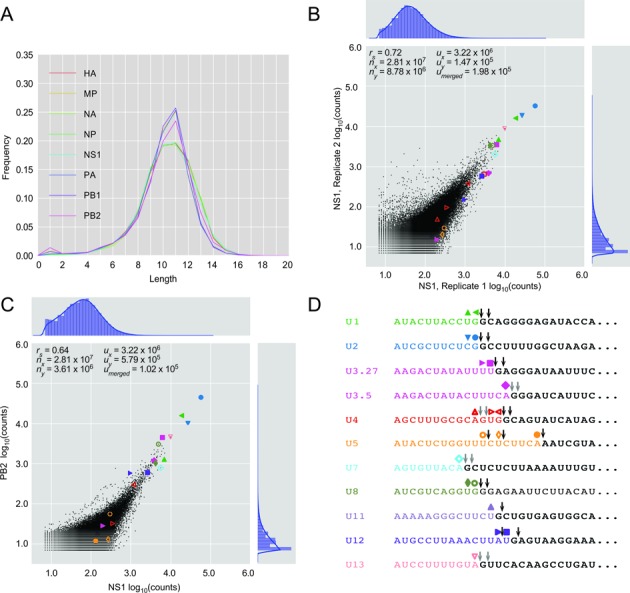
Different influenza mRNAs are prepended with similar sets of sequences that include leaders from snRNAs and snoRNAs. (A) Length distributions of host leaders after trimming nucleotides attributed to prime-and-realign. (B) Number of reads corresponding to the same host leader sequences in two biological replicates of NS1, 4 h.p.i., after trimming nucleotides attributed to prime-and-realign. Shapes indicate values for leaders mapping to the annotated 5′ ends of snRNAs and snoRNAs, colored as in panel (D). Otherwise, as in Figure 1D. (C) Number of reads corresponding to the same host leader sequences from NS1 and PB2 mRNAs, 4 h.p.i., after trimming prime-and-realigned nucleotides. Otherwise, as in (B). (D) Abundant host leaders corresponding to the annotated 5′ ends of snRNAs and abundant snoRNAs. The last nucleotide of each abundant trimmed host leader highlighted in (B) and (C) is indicated (colored shape), as is the presumed cleavage site for each of these host leaders (black arrows showing unambiguous sites or two gray arrows showing alternative cleavage sites for the same host leader).
