Figure 8.
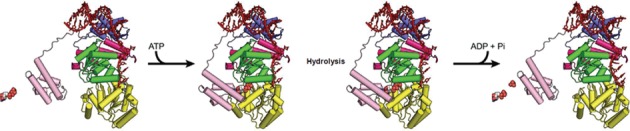
Model of the coupling of ATP hydrolysis/DNA translocation cycle with HRDC-mediated conformational change. We assume that in the absence of nucleotide the HRDC domain is disengaged, and the two RecA domains adopt a different relative orientation in which the distance between ssDNA binding motifs on D1 and D2 is approximately equal to the phosphate to phosphate distance of one nucleotide step (which in our model is based on the conformation of HsRECQ1). Nucleotide binding and hydrolysis is accompanied by the association of the HRDC domain, the adoption of the conformation of D1 and D2 found in the BLM crystal structures and the transition of the DNA from the pre-translocation state (found in our structure), to the post-translocation state based on PDB: 4O3M.
