Figure 2.
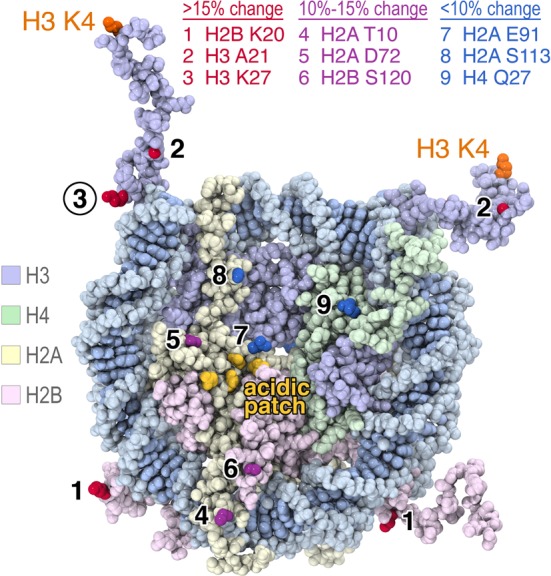
Fluorescence positions on the nucleosome affected by LSD1/CoREST binding. Oregon Green 488 was conjugated site-specifically to histones engineered with unique cysteines, reconstituted with appropriate histones and Widom 601 147 bp DNA and used in fluorescence quenching nucleosome binding experiments. The results are mapped on the space filling representation of the 1.9 Å crystal structure of the nucleosome core particle (PDB code 1KX5). Nucleosomes labeled on H2B K20, H3 A21 and H3 K27 showed >15% fluorescence quenching upon LSD1/CoREST binding (side chains shown in red), nucleosome labeled on H2A T10, H2A D72 and H2B S120 produced 10–15% fluorescence quenching (purple residues) while nucleosomes labeled on H2A E91, H2A S113 and H4 Q27 produced less than 10% fluorescence quenching (blue residues). The four H2A residues in the nucleosome acidic patch, E61, E64, D90 and E92, are shown in yellow. Figure prepared using PyMOL molecular graphics software (37).
