Figure 5.
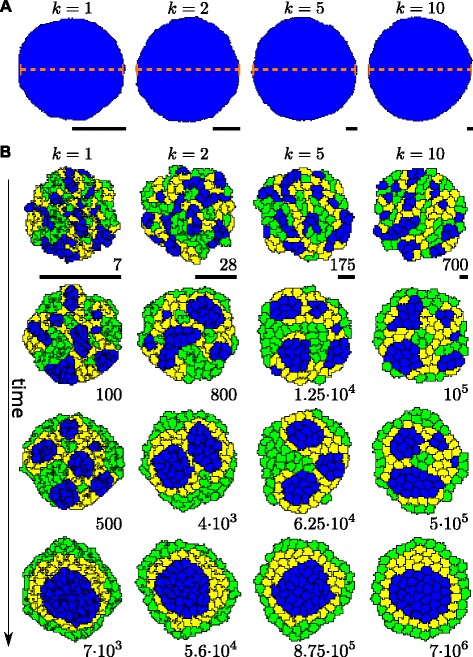
CPM dynamics at different resolutions.(A) Simulations of a single cell at different resolutions (k=1, 2, 5, 10). For visual comparison, each panel shows the simulation after 10,000 MCS, but rescaled with the k-value used for the simulation. See Appendix C for parameter details. Scale bars indicate a length of 50 lattice points. The dashed red lines indicate the analytically predicted cell diameter, a very close correspondence is found for all resolutions. (B) Simulations of a tissue containing three different cell types, G(reen), Y(ellow), and B(lue), as well as M(edium), again at four different resolutions. As before, each panel shows the simulation rescaled with the k-value used for visual comparison, with time (expressed in Monte Carlo time steps (MCS), in which each point of the lattice is considered for an update once) indicated below, and scale bars representing a length of 50 lattice points. The time required for formation and rounding up of clusters scales with k 2 (see upper row), while the time required for the drift and merging of small clusters, eventually leading to complete cell sorting, scales with k 3 (middle rows and lower row). Further parameter details are given in Appendix C.
