Figure 7. Ancient Mammalian TEs Globally Remodeled PGR Binding Site Architecture across the Genome.
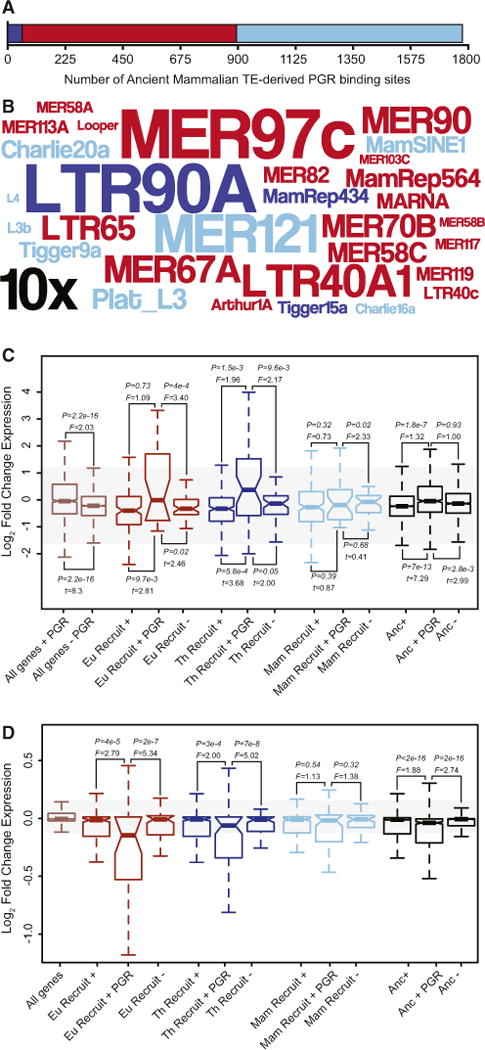
(A) The number of PGR ChIP-seq peaks that contain Mammalian-specific (light blue), Therian-specific (blue), and Eutherian-specific (red) TEs in primary cultures of hESCs treated with 10 nM 17β-estradiol, 100 nM medroxyprogesterone acetate, and 1 mM 8-bromo-cAMP.
(B) Word cloud of ancient mammalian TEs enriched within PGR ChIP-seq peaks. Colors follow (A). Inset scale (10×) shows 10-fold enrichment. Only elements with R2-fold enrichment are shown.
(C) Genes associated with ancient mammalian TE-derived PGR binding sites (+ PGR) are more strongly differentially regulated in hESCs upon decidualization than genes associated with ancient mammalian TE as a whole (+) and genes without TE-derived regulatory elements (−). Eu Recruit, Eutherian recruited gene; Th Recruit, Therian recruited gene; Mam Recruit, Mammalian recruited gene; Anc, ancestrally expressed gene. F is the ratio of variances from a two-sample F test.
(D) Genes associated with ancient mammalian TE-derived PGR biding sites (+ PGR) are more strongly dysregulated by PGR knockdown in DSCs than either genes associated with ancient mammalian TE as a whole (+) and genes without TE-derived regulatory elements (−). F is the ratio of variances from a two-sample F test.
