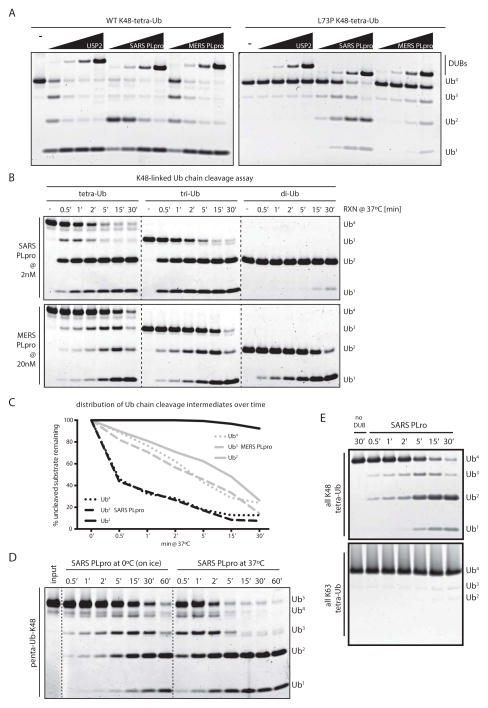
Figure 2. Enhanced activity of SARS PLpro on longer K48-linked ubiquitin chains.
(A) K48-linked wild-type and DUB-resistant (UbL73P) tetra-ubiquitin chains assayed with a 1/5-fold serial-dilution SARS PLpro, MERS PLpro and USP2CD (starting at 2μM DUB), showing how both MERS and SARS PLpro can tolerate Pro in P4 in the context of full-length ubiquitin when it is part of a tetra-ubiquitin chain. (B) K48-linked ubiquitin chains (tetra-, tri- and di-Ub) were cleaved in a time-course assay by SARS PLpro (at 2nM, top panels) and by MERS PLpro (at 20nM, bottom panels); highlighting the different cleavage patterns of the two DUBs. Dotted lines indicate cropped images from three gels, ran side by side. (C) Quantification of cleavage rates by SARS and MERS PLpro from (B). (D) K48-linked penta-ubiquitin chains were cleaved in a time-course assay by SARS PLpro (20nM) at 0°C (on ice) and 37°C, and visualized by SDS-PAGE and SYPRO-staining. Dotted lines are included for clarity. (E) Comparison of tetra-Ub cleavage by SARS PLpro of K48- versus K63-linked chains.