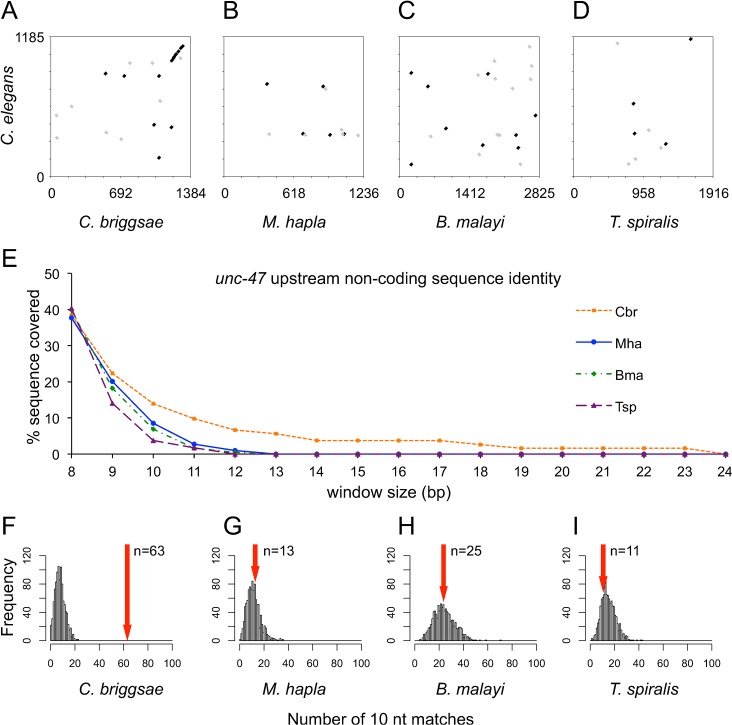
Fig 6. Sequence similarity of orthologous unc-47 cis elements.
(A-D) Dotplots showing perfect sequence identity (in 10 nucleotide windows) of the unc-47 upstream sequences in direct (black) and reverse (gray) orientations between C. elegans and (A) C. briggsae, (B) M. hapla, (C) B. malayi, (D) T. spiralis. (E) Percentage (y-axis) of unc-47 upstream sequences composed of short blocks of sequence identity with C. elegans across various window sizes (x-axis). (F-I) Distributions of the number of perfect 10 nucleotide matches between 1000 replicates of reshuffled C. elegans unc-47 upstream sequence and the upstream unc-47 sequences of (F) C. briggsae, (G) M. hapla, (H) B. malayi, (I) T. spiralis. Trinucleotide frequencies of the original C. elegans unc-47 upstream sequence were retained. Red arrows indicate the actual number of 10 nucleotide matches between C. elegans and each ortholog. Only C. briggsae has more than would be expected by chance (see Results). Note that B. malayi sequence is approximately twice as long as that of C. elegans, C. briggsae, and M. hapla.