Figure 4. In silico prediction of A. gambiae aga-miR-305 targets using RNAHybrid, miRanda, and PITA.
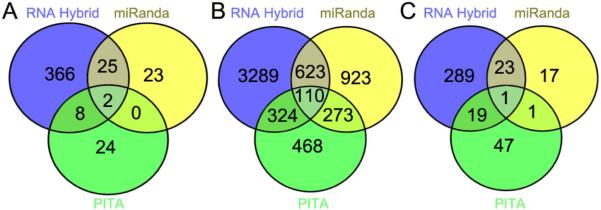
Gene targets of aga-miR-305 were predicted within the (A) 5’ untranslated region, (B) coding sequence, and (C) 3’ untranslated region of A. gambiae genes (Vectorbase: AgamP.37) using the software programs RNAHybrid, miRanda, and PITA. Details of the parameters used for each program are provided in Materials and Methods. Venn diagrams were created using the online software Venny (Oliveros, (2007, accessed June 2014)).
