Summary
Iron availability is a key determinant of virulence in the pathogenic fungus Cryptococcus neoformans. Previous work revealed that the ESCRT (endosomal sorting complex required for transport) protein Vps23 functions in iron acquisition, capsule formation and virulence. Here, we further characterized the ESCRT machinery to demonstrate that defects in the ESCRT-II and III complexes caused reduced capsule attachment, impaired growth on haem and resistance to non-iron metalloprotoporphyrins. The ESCRT mutants shared several phenotypes with a mutant lacking the pH-response regulator Rim101 and, in other fungi, the ESCRT machinery is known to activate Rim101 via proteolytic cleavage. We therefore expressed a truncated and activated version of Rim101 in the ESCRT mutants and found that this allele restored capsule formation but not growth on haem, thus suggesting a Rim101-independent contribution to haem uptake. We also demonstrated that the ESCRT machinery acts downstream of the cAMP/protein kinase A pathway to influence capsule elaboration. Defects in the ESCRT components also attenuated virulence in macrophage survival assays and a mouse model of cryptococcosis to a greater extent than reported for loss of Rim101. Overall, these results indicate that the ESCRT complexes function in capsule elaboration, haem uptake and virulence via Rim101-dependent and independent mechanisms.
Introduction
Iron is a key factor in the virulence of Cryptococcus neoformans, a pathogenic fungus that causes life-threatening meningoencephalitis in immunocompromised individuals. This is because of its role as a cofactor in critical biochemical functions and its influence on the elaboration of two major virulence factors, the polysaccharide capsule and melanin. As in other pathogens, C. neoformans has developed multiple strategies to acquire iron from vertebrate hosts, which actively limit the iron available to invading pathogens through nutritional immunity (Cassat and Skaar, 2013). A key strategy for the fungus is the use of a high-affinity iron uptake system comprised of the iron permease Cft1 and the ferroxidase Cfo1 (Jung et al., 2008; 2009); these proteins are required for full virulence in a murine inhalation model of cryptococcosis and for iron acquisition from transferrin. Additionally, C. neoformans deploys a number of siderophore transporters although only one of these has been tested and found not to play a role in virulence (Tangen et al., 2007). C. neoformans can also acquire iron from haem and from haem-containing proteins such as haemoglobin, the most abundant iron sources in vertebrate hosts (Hu et al., 2013). We recently identified the extracellular mannoprotein Cig1 as a potential haem-binding protein that functions in C. neoformans to acquire iron from haem (Cadieux et al., 2013). Deletion of CIG1 resulted in delayed growth on haem at neutral pH, reduced susceptibility to non-iron metalloprotoporphyrins (MPs) that require a haem uptake system for toxicity, and attenuated virulence in a strain that also lacked high affinity iron uptake. Additionally, a candidate ferric reductase participates in iron acquisition from haem and in virulence (Saikia et al., 2014).
A number of transcription factors regulate the expression of iron acquisition functions in C. neoformans and these include Cir1, HapX and Rim101 (O'Meara et al., 2010; Kronstad et al., 2013). Remarkably, Cir1 regulates many of the iron uptake systems and all of the known major virulence factors (Jung et al., 2006). HapX is required for growth on haem and is a major regulator of iron-dependent metabolic functions such as respiration (Jung et al., 2010). The pH-responsive factor Rim101 regulates iron uptake functions including the expression of Cig1, and a rim101 mutant cannot use iron from haem due to a defect in uptake (O'Meara et al., 2010; Cadieux et al., 2013). Rim101 mutants are also defective in cell-associated capsule, and they display extensive shedding of capsule polysaccharide as well as hyper-activation of the host immune response due to cell surface changes (O'Meara et al., 2010; 2013).
In other fungi, Rim101 is activated by proteolytic processing in which the D/E rich C-terminal tail of the protein is cleaved by the calpain-like protease Rim13 to yield an active, N-terminal polypeptide containing three zinc finger motifs (Li and Mitchell, 1997; Mingot et al., 1999; Xu et al., 2004; Wolf et al., 2010; Wolf and Davis, 2010). Additional proteins involved in the processing include Rim20, a regulatory component, which binds to the YPXL/I sites within the D/E rich domain of Rim101 (Xu and Mitchell, 2001; Vincent et al., 2003). In some fungi, for example Saccharomyces cerevisiae and Candida albicans, Rim20 contains a Bro1 domain, which interacts with Vps32 (also called Snf7) thereby establishing a connection to the endosomal sorting complex required for transport (ESCRT) machinery (Ito et al., 2001; Bowerset al., 2004; Xu et al., 2004; Boysen and Mitchell, 2006). In particular, the ESCRT-I, II and III complexes are part of the signaling pathway that results in activation of Rim101 by proteolytic processing in response to alkaline pH (Xu et al., 2004; Penalva et al., 2008). It is also notable that Rim101 activation is mediated by the cAMP/protein kinase A pathway in C. neoformans (O'Meara et al., 2010).
We previously constructed and screened a collection of 30,000 Agrobacterium-mediated insertion mutants for defects in growth on haem. Among the mutants, we identified and characterized a role for the ESCRT-I component, Vps23, in haem uptake and utilization, and also in capsule formation and virulence (Hu et al., 2013). Interestingly, this mutant shared the first two phenotypes with a mutant defective in Rim101. The ESCRT machinery is composed of several protein complexes (ESCRT-0, -I, -II, -III) and functions in the targeting of monoubiquitinated membrane proteins to the vacuole for degradation (Hurley and Emr, 2006). The ESCRT machinery also governs the biogenesis of multivesicular bodies (MVB) and has roles in cytokinesis and the budding of enveloped viruses at the plasma membrane (Hanson et al., 2009; Wollert et al., 2009; Hurley, 2010; Hurley and Hanson, 2010; Henne et al., 2011; Morita et al., 2011).
As mentioned, the ESCRT pathway participates in the activation of Rim101 in fungi and it is possible, therefore, that the haem defect of the vps23 ESCRT-I mutant is due, at least in part, to the inability to activate Rim101. In general, several unanswered questioned remained from the study of Vps23, especially given the connections outlined above between ESCRT complexes, Rim101, Cig1, PKA and the use of haem. For example, are all of the ESCRT complexes or only a subset involved in haem uptake and capsule elaboration? In addition to the information for VPS23, it is known that disruption of VPS25, a component of the ESCRT-II complex, causes sensitivity to copper or iron depletion, and a reduction in capsule size (Chun and Madhani, 2010). Similarly, does the influence of ESCRT on these processes occur completely through activation of Rim101 or does the complex function in other pathways? Finally, does the ESCRT pathway interact with the cAMP/PKA pathway in the regulation of capsule size and is there an additional connection with iron uptake? To address these questions, we characterized the roles of components representing each of the ESCRT complexes. We discovered that the ESCRT complexes have a conserved role in C. neoformans, relative to other fungi, and that the mutants lacking ESCRT-II and III components shared the phenotypes of the vps23 (ESCRT-I) mutant. However, mutants missing accessory components behaved like the wild-type (WT) strain in terms of capsule formation and haem use, while a vps27 (ESCRT-0) mutant had a subtle growth defect on haem. The phenotypes of the ESCRT-I, II and III mutants in capsule formation were largely explained by the interaction with the Rim101 pathway. However, Rim101-independent roles for haem use and virulence were also identified, and the functions of the ESCRT complexes only partially contribute to the influence of PKA activation on capsule size.
Results
Identification and mutation of the core components of ESCRT machinery
Our goal in this study was to examine the role of representatives of each ESCRT complex in iron acquisition from haem, capsule formation and virulence, and to investigate the extent to which ESCRT functions were dependent on activation of Rim101. Initially, the genome sequence was examined to identify the C. neoformans orthologs of ESCRT complex proteins as characterized in other fungi. This analysis identified most of the known proteins in the ESCRT complexes with the exception of two ESCRT-I components (Vps37 and Mvb12) (Table 1). We then generated deletion mutations in selected genes encoding representatives of each ESCRT complex including VPS27 (ESCRT-0), VPS22 (ESCRT-II), VPS20 and SNF7 (ESCRT-III) (Table 1). We also mutated the VPS4 gene encoding an AAA-ATPase and the BRO1 gene encoding an accessory protein that interacts with Snf7. The mutations for each gene were generated with two different resistant marker cassettes (i.e., neomycin and hygromycin) to obtain independent mutants, and the deletions were confirmed by PCR and Southern hybridization (data not shown). Two independent mutants for each gene were subsequently used in all phenotypic analyses, and the data are presented for one representative mutant. Note that the previously characterized mutants in VPS23 (ESCRT-I) are included throughout this study for comparison (Hu et al., 2013).
Table 1.
Components of the ESCRT machinery in C. neoformans var. grubii
| Complex | Protein name | Locus tag | Accession ID | Saccharomyces cerevisiae protein |
|---|---|---|---|---|
| ESCRT-0 | Vps27 | CNAG_02167 | AFR95653.2 | YNR006W |
| Hse1 | CNAG_05882 | AGV14503.1 | YHL002W | |
| ESCRT-I | Vps23 | CNAG_01720 | AFR97923.1 | YCL008C |
| Vps37 | unidentified | YLR119W | ||
| Vps28 | CNAG_00676 | AFR92805 | YPL065W | |
| Mvb12 | unidentified | YGR206W | ||
| ESCRT-II | Vps22 | CNAG_05704 | AFR96022.1 | YPL002C |
| Vps25 | CNAG_04863 | AFR97353.1 | YJR102C | |
| Vps36 | CNAG_00248 | AFR92381.1 | YLR417W | |
| ESCRT-III | Vps2 | CNAG_05478 | AFR98910.2 | YKL002W |
| Vps20 | CNAG_01265 | AFR94877.1 | YMR077C | |
| Vps24 | CNAG_00421 | AFR92554.2 | YKL041W | |
| Snf7 (Vps32) | CNAG_01583 | AFR97788.1 | YLR025W | |
| Vps60 | CNAG_05715 | AFR96033.1 | YDR486C | |
| Did2 (Vps46) | CNAG_00470 | AFR92602.1 | YKR035W-A | |
| Ist1 | CNAG_02535 | AFR95298.1 | YNL265C | |
| ESCRT-DS | Vps4 | CNAG_04510 | AFR97229.1 | YPR173C |
| Bro1 (Vps31) | CNAG_03372 | AFR96592.1 | YPL084W | |
| Doa4 | CNAG_00757 | AFR92887.2 | YDR069C | |
| Vta1 | CNAG_05747 | AGV14481.1 | YLR181C |
The genes for the proteins indicated in bold were characterized in this study.
We first tested the ESCRT mutants for defects in the conserved function of endosomal trafficking by using the lipophilic dyes FM4-64 and MDC64 to observe internal membrane structures. The cells of the WT strain demonstrated strong staining of the vacuolar membrane, with little or no staining in the cytoplasm. Cells of the vps22, vps23, vps27, vps20 and snf7 mutants showed accumulations of perivacuolar staining, indicating the presence of so-called class E-like exclusion bodies, as reported for ESCRT mutants in other fungi (Supplemental Fig. S1A, Bryant and Stevens, 1998; Hu et al., 2013). In contrast, the cells of the vps4 and bro1 mutants displayed only minor differences from WT. We also tested the strains for susceptibility to brefeldin A (BFA), a drug that arrests the anterograde transport of proteins between the ER and the Golgi compartments. The vps22, vps27, snf7 and vps20 mutants displayed increased susceptibility to BFA, as was previously found with the vps23 mutant (Supplemental Fig. S1B; Hu et al., 2013). The bro1 and vps4 mutants were similar to the WT in susceptibility. These results suggest that the ESCRT–0, I, II, and III complexes, but not the accessory proteins, are involved in intracellular trafficking (e.g., endocytosis and ER-Golgi transport). Taken together, we conclude that the ESCRT components identified in C. neoformans make similar contributions to endosomal trafficking as found in S. cerevisiae, C. albicans and other fungi (Bryant and Stevens, 1998; Xu et al., 2004).
Loss of ESCRT-I, -II and –III components causes a growth defect on haem
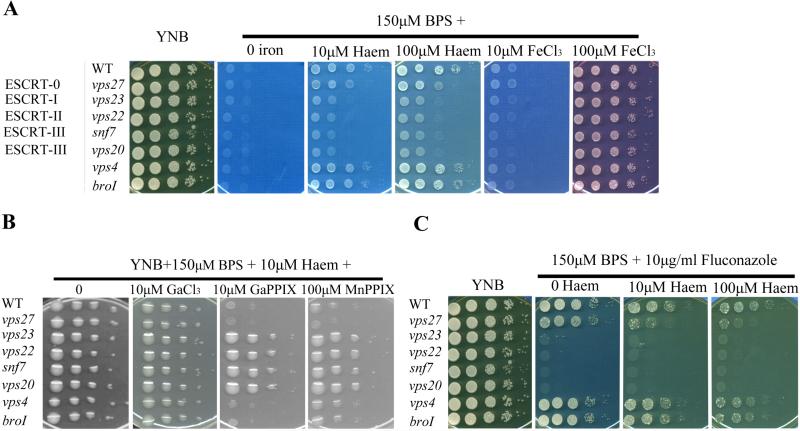
We next examined the contributions of the representative ESCRT components to growth on various iron sources. The strains were first grown for two days in YNB-LIM medium (YNB containing the iron chelator bathophenanthroline disulfonate (BPS)) to exhaust intracellular iron stores, and growth was then tested in spot assays on YNB-LIM at neutral pH (pH 7.0), without or with FeCl3 or haem (Fig. 1A). The WT strain and all ESCRT mutants grew robustly on iron-replete YNB medium, but failed to grow on iron-depleted medium (YNB-LIM). The WT strain also grew on YNB-LIM with the addition of 10 μM FeCl3 and all other iron sources. Similarly, all of the ESCRT mutants (vps27, vps23, vps22, vps20, snf7, vps4 and bro1) generally grew like WT on medium supplemented with either 10 μM or 100 μM FeCl3, although the growth on the medium supplemented with 10 μM FeCl3 was limited. However, as previous found for the vps23 mutant (Hu et al., 2013), the deletion mutants lacking ESCRT-II (Vps22) and ESCRT-III (Snf7 and Vps20) proteins showed reduced growth on YNB-LIM supplemented with haem at either 10 μM or 100 μM at pH 7.0. The mutants in VPS27 (ESCRT-0) demonstrated a subtle (partial) growth defect in the presence of haem at either 10 μM or 100 μM at pH 7.0. However, the deletion mutants for the two accessory ESCRT components (vps4 and bro1) grew like WT on the low iron medium supplemented with either haem or FeCl3 (at either 10 μM or 100 μM) (Fig. 1A). Growth assays were also performed in liquid YNB-LIM for these deletion strains, with similar results (Supplemental Fig. S2). Overall, we found that the ESCRT-I (Vps23), ESCRT-II (Vps22) and ESCRT–III (Snf7 and Vps20) proteins, and to a lesser extent ESCRT-0 (Vps27), but not Bro1 and Vps4, are required for efficient iron acquisition from haem.
Figure 1. The ESCRT machinery is required for growth on haem and haem uptake.
A. The growth of ESCRT mutants and the WT strain was tested on YNB and YNB plus BPS supplemented with either haem or FeCl3 at 10 μM or 100 μM at pH 7.0.
B. Susceptibility to non-iron metalloporphyrins was tested by spotting cells onto low-iron medium plus 10 μM haem in the absence or presence of 10 μM gallium protoporphyrin (Ga-PPIX), 100 μM manganese protoporphyrin (Mn-PPIX), or 10 μM GaCl3 as indicated.
C. Suppression of the fluconazole susceptibility of the ESCRT-0, -I, -II and –III mutants was tested by addition of exogenous haem at 0, 10 μM or 100 μM to YNB plus BPS medium containing 10 μg ml−1 fluconazole.
For all assays, the strains were grown in low-iron medium and ten-fold serial dilutions of were spotted on agar plates, with subsequent incubation for 2-3 days at 30°C.
Loss of ESCRT-I, -II and –III proteins reduces susceptibility to non-iron metalloprotoporphyrins
The non-iron MPs gallium protoporphyrin (Ga-PPIX) and manganese protoporphyrin (Mn-PPIX) are haem analogs that enter cells via haem uptake pathways to cause toxicity (Stojiljkovic et al., 1999). We previously demonstrated that defects in components of the haem uptake system (i.e., Cig1, Rim101 and Vps23) cause reduced susceptibility to MPs (Cadieux et al., 2013, Saikia et al., 2014, Hu et al., 2013). We therefore predicted that the ESCRT-II and III complexes would also influence susceptibility to MPs if they participated in haem uptake. To test this hypothesis, we compared the growth of the WT strain and all of the ESCRT mutants on haem in the presence or absence of Ga-PPIX or Mn-PPIX, after the strains had been iron starved. All of the strains grew well in the absence or presence GaCl3 on the defined low iron medium (LIM) with added haem indicating that gallium does not have toxic effect on the strains (Fig. 1B). Deletion of VPS23 (as previously tested by Hu et al., 2013), VPS22, VPS20 and SNF7 resulted in reduced susceptibility to both GaPPIX and MnPPIX. However, the vps27, vps4 and bro1 mutants each grew as well as WT on LIM supplemented with either GaPPIX or MnPPIX indicating that these proteins are dispensable for haem uptake. Overall, these data support the hypothesis that the ESCRT-I, II and III complexes contribute to haem uptake by C. neoformans.
Haem does not restore the growth of ESCRT mutants on fluconazole
Haem plays an important role in ergosterol biosynthesis because some of the enzymes in the pathway are haem dependent, including lanosterol 14-α-demethylase (Crisp et al., 2003; Jung et al., 2009; Kim et al., 2012). In this context, we speculated that ESCRT mutants with defects in haem uptake would show increased susceptibility to the azole drugs that target ergosterol biosynthesis. Indeed, we found that loss of VPS23 (−I), VPS22 (−II) and VPS20 and SNF7 (−III) caused a marked increase in susceptibility to fluconazole (Fig. 1C). It is possible that loss of ESCRT function may generally compromise membrane integrity and thus exacerbate the impact of reduced ergosterol due to fluconazole inhibition of lanosterol 14-α-demethylase. ESCRT functions may also play a role in haem uptake and/or in trafficking haem to the endoplasmic reticulum to support ergosterol biosynthesis. To begin to investigate these possibilities, we tested the influence of exogenous haem on drug susceptibility and found that addition of either 10 μM or 100 μM failed to restore the growth of the ESCRT mutants in the presence of fluconazole (Fig. 1C; Supplemental Fig. S3). In contrast, the deletion of VPS4 or BRO1 had no influence on drug susceptibility. However, deletion of VPS27 caused a slight increase in susceptibility.
The results for the ESCRT mutants are in contrast to our previous demonstration that deletion of the CFO1 gene encoding the ferroxidase component of high-affinity iron trafficking complex resulted in an increased susceptibility to fluconazole that could be mitigated by exogenous haem (Jung et al., 2009). The loss of Cfo1 reduces iron acquisition but does not influence haem uptake (Jung et al., 2009). A comparison of the influence of haem on fluconazole susceptibility for the cfo1, vps23, rim101 and vps23cfo1mutants revealed that loss of Vps23 function blocked the ability of exogenous haem to ablate the fluconazole hyper-susceptibility of the cfo1 mutant (Supplemental Fig. S3). We hypothesize that haem restores the growth of the cfo1 mutant on fluconazole because an intact uptake pathway supplies haem for ergosterol biosynthesis (Jung et al., 2009, Kim et al., 2012). In this context, our findings are consistent with the possibility that the ESCRT–I, -II and –III complexes, but not the accessory components Vps4 and Bro1, participate in haem trafficking. However, a broader impact on membrane composition should be examined because we found that defects in ESCRT–I, -II and –III functions also caused increased susceptibility to the polyene antifungal drug amphotericin B which binds ergosterol in the plasma membrane (data not shown).
The ESCRT machinery influences capsule and melanin formation
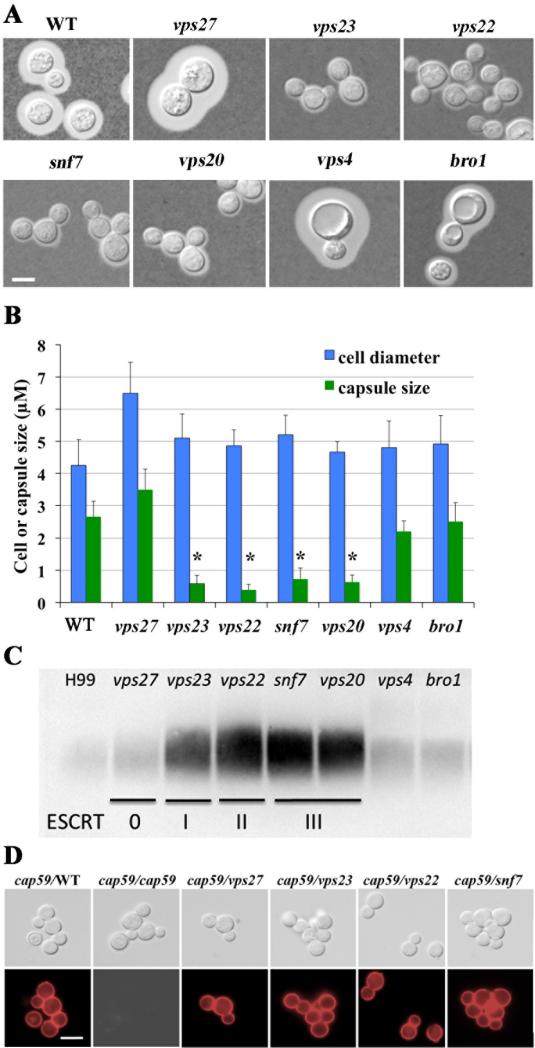
Loss of the ESCRT-I protein Vps23 reduces attachment of capsule polysaccharide to the cell wall and melanin formation (Hu et al., 2013), and we therefore tested whether mutants in other ESCRT components shared these phenotypes. Initially, the ESCRT mutants were grown in defined LIM and cell-associated capsule was observed by staining with India ink. As expected, the WT cells produced a large capsule while deletion of VPS23, VPS22, VPS20 and SNF7 caused a marked reduction in capsule size (Fig. 2A). Measurements of capsule diameter revealed that the reduction in the vps23, vps22, vps20 and snf7 mutant strains versus WT was statistically significant (Fig. 2B). These observations were confirmed with independent mutants for each gene (data not shown). In contrast, deletion of VPS27, VPS4 and BRO1 did not influence capsule size compared with the WT strain (Figs. 2A and 2B). We conclude that the ESCRT-I, II and –III complexes are involved in the elaboration of cell-associated capsule.
Figure 2. Altered capsule size and polysaccharide shedding for the ESCRT mutants.
A. Cells were grown in defined low iron medium at 30°C for 48 h, and capsule formation was assessed by India ink staining for the indicated strains. Bar = 5 μm.
B. Fifty cells of each strain from the assays in A were measured to determine the cell diameter and capsule radius. Each bar represents the average of the 50 measurements with standard deviations. Statistical significance relative to the WT capsule size is indicated by an asterisk (Student t test (P<0.05)).
C. The electrophoretic mobility and relative quantities of shed polysaccharide were assessed by immunoblotting with the anti-GXM antibody mAb 18b7 to detect the capsule. The DIC images are shown on the top row and the immunofluorescence images are shown on the bottom row.
D. A capsule transfer assay to determine whether polysaccharide secreted from cells of the WT strain or the ESCRT mutants into conditioned medium (CM) could attach to cells of an acapsular cap59 acceptor strain. CM from the cap59 mutant was used as a negative control because this strain does not shed capsule polysaccharide. Cells were labeled with the anti-GXM antibody mAb 18b7 to detect capsule, and Alexa Fluor anti-mouse IgG (secondary antibody) was used for immunofluorescence. Bar = 10 μm.
We next used immunoblotting of culture supernatants to examine shedding of the major polysaccharide glucuronoxylomannan (GXM) as an indicator of a defect in capsule attachment to the cell wall, as previously seen with the vps23 mutant (Hu et al., 2013). As shown in figure 2C, the vps22, vps20 and snf7 mutants, along with the vps23 mutant, shed more GXM than the WT strain. In contrast, the vps27, vps4 and bro1 mutants had similar levels of GXM in the culture supernatant as the WT strain (Fig. 2C). We further established that the ESCRT machinery actually influences capsular attachment rather than capsule biosynthesis by a capsule transfer assay (Reese and Doering, 2003; Fang et al., 2012). In this assay, conditioned medium (CM) was prepared as a source of capsule polysaccharide by growing the WT strain and the ESCRT mutants in defined LIM to induce capsule for 3 days, and CM from acapsular cap59 cells was used as a negative control. Indirect immunofluorescence microscopy revealed that the acapsular cap59 cells could attach polysaccharide from the CM medium of the vps27, vps23, vp22, snf7 and vps20 mutants (Fig. 2D). These data suggest that the capsule from the mutants lacking components of the ESCRT-I, II or –III machinery was competent (at least in part) for attachment. The small capsule phenotype for these mutants is therefore likely due to a defect in capsule attachment rather than defective GXM synthesis. A change in the cell wall to influence attachment is consistent with our finding that the ESCRT-I, II and III mutants show increased susceptibility to agents such as congo red and caffeine that challenge cell wall integrity (Supplemental Fig. S4).
Deletion of the core components of the ESCRT machinery (vps23, vp22, snf7 and vps20) also caused reduced melanin production (Supplemental Fig. S5). In particular, the snf7 mutant cells displayed the greatest reduction in melanin on the L-DOPA medium, while deletion of VPS27, VPS4 and BRO1 did not influence melanin production. We also noted that the snf7 mutant strain grew slower than the other mutants or the WT strain at 37°C, potentially indicating a distinct role of the Snf7 protein (Supplemental Fig. S5). Overall, these data indicate that the ESCRT machinery also influence melanin formation, perhaps through an influence on intracellular trafficking of laccase.
ESCRT mutants share phenotypes with a rim101 mutant, and Snf7 interacts with Rim20 in the Rim101 activation pathway
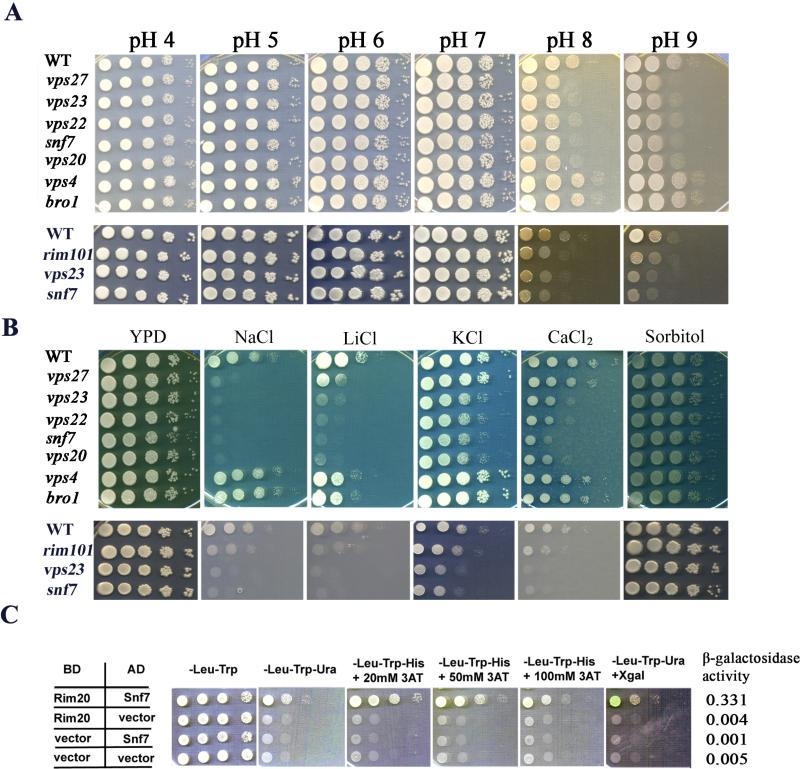
As mentioned, Rim101 regulates the response to pH in fungi and activation of the protein is dependent on ESCRT functions (Ito et al., 2001; Bowers et al., 2004; Xu et al., 2004; Boysen and Mitchell, 2006; O'Meara et al., 2010). To determine whether the defects in capsule attachment and growth on haem for the ESCRT mutants were due to failure to activate Rim101, we first tested the mutants for phenotypes seen in the rim101 mutant including impaired growth at alkaline pH and in the presence of NaCl and LiCl (O'Meara et al., 2010). We found that all of the strains grew equally well on YPD at pH 4 to pH 7 (Fig. 3A). However, the vps27, vps23, vps22, snf7 and vps20 mutants demonstrated reduced growth compared with the WT strain on YPD at both pH 8 and pH 9 indicating that these ESCRT components (0, I, II, and III) were required for the robust growth at alkaline pH (Fig. 3A). The impaired growth of a rim101 mutant at alkaline pH is also shown for comparison. In contrast, deletion of the VPS4 and BRO1 genes did not influence growth at alkaline pH (Fig. 3A). Moreover, the vps23, vps22, snf7 and vps20 mutants showed reduced growth on YPD medium supplemented with either 1.5M NaCl, 200 mM LiCl, 1.5M KCl or 250 mM CaCl2 (Fig. 3B). These mutants grew at the WT level on YPD with 1.5M sorbitol, thus indicating that salt rather than osmotic stress caused their growth defects. The vps27 mutant showed defects only on NaCl and LiCl, and the vps4 and bro1 mutants grew like the WT strain under all salt stress conditions (Fig. 3B). Taken together, these results indicate that the ESCRT-0, I, II and III mutants share phenotypes with a rim101 mutant.
Figure 3. Susceptibility of ESCRT mutants to alkaline pH and salt stress, and interactions with Rim20 in the Rim101 pathway.
A. The vps27, vps23, vps22, vps20 and snf7 mutants had impaired growth at alkaline pH. Cells of ten-fold serial dilutions of the indicated strains were spotted onto buffered YNB plates at the different pH, and the plates were incubated at 30°C for 2 days before being photographed. The vps4 and bro1 mutants grew like the WT strain in all media.
B. The vps27, vps23, vps22, vps20 and snf7 mutants showed increased susceptibility to salt stress. Ten-fold serial dilutions of cells of the indicated strains were spotted onto solid YPD without or with 1.5 M NaCl, 1.5 M KCl, 250 mM CaCl2, 1.5 M sorbitol, or 200 mM LiCl. The plates were incubated at 30°C for the following times: sorbitol, 3 days; CaCl2, 3 days; KCl, 5 days; NaCl, 5 days; LiCl, 10 days. The vps4 and bro1 mutants grew like the WT strain in all media.
C. Snf7 physically interacts with the regulatory component of Rim101 pathway, Rim20, as determined by the yeast two-hybrid assay. BD and AD indicate the GAL4 binding and activation domains fused to Rim20 and Snf7, respectively. The vector designation indicates the empty vector control. All combinations of transformants grew in the absence of leucine (Leu) and tryptophan (Trp) confirming plasmid retention in the strains. Only yeast cells transformed with plasmids containing SNF7 and RIM20 grew in the absence of uracil (Ura) or histidine (His), confirming an interaction to allow expression of URA3 and HIS3. 3-amino-1,2,4-trazole (3AT) was included at different concentrations a part of the detection of HIS3 expression. Quantitative analysis of β-galactosidase activity was performed using ONPG as a substrate.
The interaction between the ESCRT machinery and the Rim101 pathways has not yet been demonstrated in C. neoformans. We therefore employed a yeast two-hybrid assay to demonstrate that the ESCRT–III protein Snf7 physically interacts with Rim20, a regulatory component of the Rim101 pathway in C. neoformans and other fungi (Li and Mitchell, 1997; Vincent et al., 2003; Xu et al., 2004; O'Meara et al., 2010). The cDNAs for Snf7 and Rim20 were fused to the GAL4 binding (AD) and activation (BD) domains, respectively, and yeast cells expressing both AD-Snf7 and BD-Rim20 were found to exhibit elevated β-galactosidase activity and grow in the absence of uracil and histidine thereby indicating an interaction (Fig. 3C). In general, this assay supports the conclusion that the ESCRT machinery in C. neoformans interacts with Rim20 to activate Rim101, as seen in other fungi (Mingot et al., 1999; Xu et al., 2004; Wolf et al., 2010; Wolf and Davis, 2010). Presumably, the absence of this interaction due to mutations in SNF7 and other ESCRT components leads to the shared phenotypes of the ESCRT and rim101 mutants.
The ESCRT machinery functions in the Rim101 pathway to regulate capsule elaboration
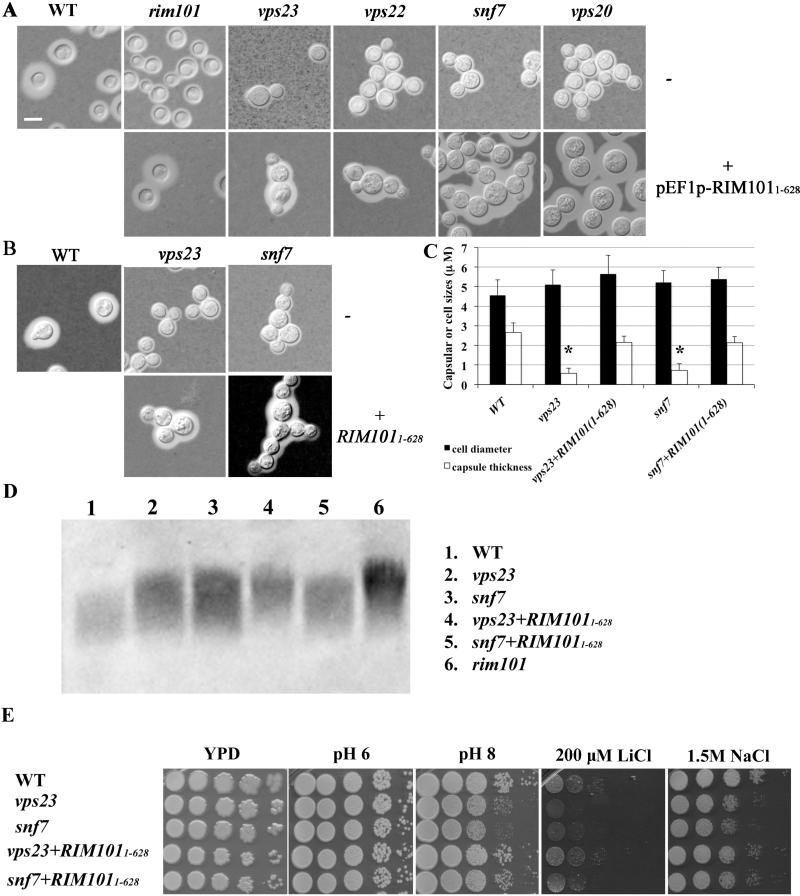
In other fungi, expression of the N-terminal portion of Rim101 corresponding to the activated form suppresses the phenotypes of a rim101 mutant at alkaline pH (Blanchin-Rolandet al., 2005; Cornet et al., 2005; Wolf et al., 2010). To further investigate whether the phenotypes in ESCRT-I, -II and –III mutants are due to the loss of Rim101 processing, we overexpressed the N-terminal region (allele designation RIM1011-628) in each of the ESCRT mutants and examined the rim101-related phenotypes. The RIM1011-628 allele lacks the D/E rich C-terminal region from amino acids 628 to 916 and encodes a predicted Rim101 polypeptide of ~ 68 kD. This allele was introduced into the WT strain and the rim101 and ESCRT mutants under control of the constitutive and highly expressed EF1 (elongation factor 1) promoter (Hu et al., 2013). We assessed capsule size in these strains by staining with India ink, and found that ectopic overexpression of the N-terminal fragment of Rim101 almost completely restored capsule formation in the rim101, vps23, vps22, snf7 and vps20 mutants (in defined LIM) (Fig. 4A). However, the growth defects at alkaline pH (pH 8-9) in these strains were only partially restored, while the growth defects in NaCl and LiCl in the strains were not altered (data not shown). The lack of complementation may be related to the reported interference of the wild-type gene with the function of the truncated Rim101 polypeptide (Zhao et al., 2013).
Figure 4. Rescue of capsule formation and the other Rim101-dependent phenotypes in ESCRT mutants by expression of an N-terminal portion of Rim101.
A. Overexpression of the N-terminal region of Rim101 from the RIM1011-628 allele restores capsule formation in a rim101 mutant and in the mutants lacking components of the ESCRT-I, -II and –III complexes. The strains also contain the WT allele of RIM101. Cells of the indicated strains were incubated in defined low-iron medium overnight at 30°C, and then observed under DIC after Indian ink staining.
B. Expression of the N-terminal region of the RIM1011-628 allele from the native promoter at the RIM101 locus rescues capsule formation for the vps23 and snf7 mutants. Cells of the indicated strains were incubated in defined low-iron medium overnight at 30°C, and then observed under DIC after Indian ink staining.
C. Fifty cells of each strain from B were measured to determine the cell diameter and capsule thickness. Each bar represents the average of the measurements with standard deviations. An asterisk indicates statistical significance by the Student t test (P<0.05) relative to the WT strain and the strains expressing RIM1011-628.
D. The electrophoretic mobility and quantity of shed polysaccharide were assessed using an anti-GXM antibody to detect the capsule.
E. Expression of the N-terminal region of the RIM1011-628 allele from the native promoter at the RIM101 locus partially restores the growth defects of the vps23 and snf7 mutants at alkaline pH and in response to salt stress. Cells of ten-fold serial dilutions of the indicated strains were spotted onto YNB plates at the different pH or in the presence of 1.5M NaCl or 200 mM LiCl, and the plates were incubated at 30°C for 2 days (pH) or 6 days (salt stress) before being photographed.
To further assess the influence of Rim101 on capsule elaboration, we replaced the wild-type RIM101 allele in the vps23 and snf7 mutants with the RIM1011-628 allele under the control of the native promoter. The RIM1011-628 allele rescued the defects in capsule formation in the vps23 and snf7 mutants (Fig. 4B). Measurement of >50 cells in each strain revealed that capsule size in either the vps23 or the snf7 strain background was approximately the same size as in WT (Fig. 4C). A capsule shedding assay further showed that capsular material in the supernatants was reduced to nearly the WT level in the strains with the RIM1011-628 allele compared to the original vps23 and snf7 mutants (Fig. 4D). This result and the restoration of capsule size suggest increased attachment of capsular material to the cell wall. Lastly, we examined the growth of the strains in alkaline conditions or under salt stress by plate spot assays. Both the vps23 and snf7 strains with the RIM1011-628 allele grew at a level similar to the WT on YPD at pH 8 or with the addition of either LiCl or NaCl (Fig. 4E). An examination of the vacuolar morphology using the lipophilic dyes FM4-64 and MDC64 with the vps23 and snf7 mutants carrying the RIM1011-628 allele revealed the presence of E-like exclusion bodies, a typical feature of ESCRT mutants. Thus the expression of the RIM1011-628 allele in these mutants did not rescue the defects of endosomal trafficking of ESCRT mutants (Supplemental Fig. S1; data not shown). In summary, the ESCRT machinery plays a role upstream of the Rim101 pathway in C. neoformans to influence the attachment of capsule material, in addition to the conserved role in the response to pH.
Defective ESCRT machinery disrupts the regulatory role of cAMP signaling on capsule elaboration
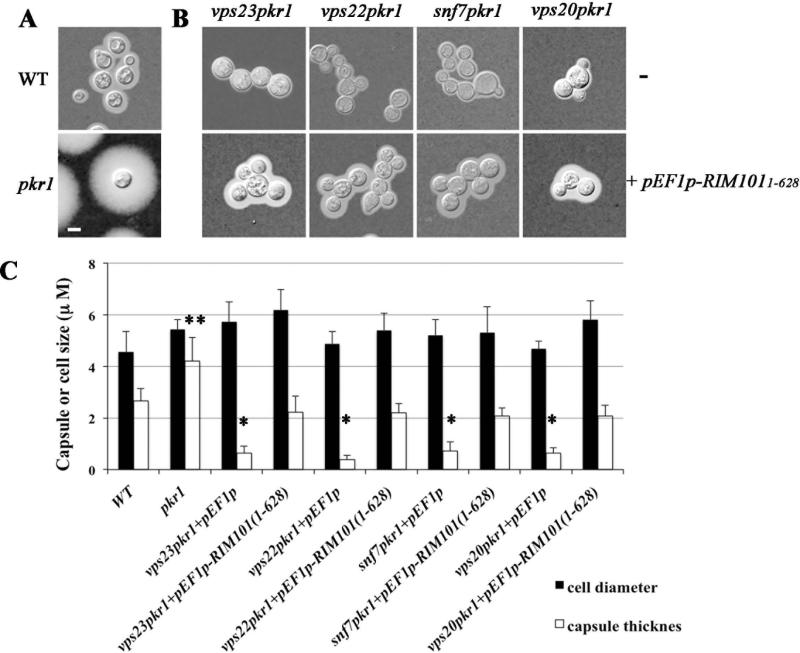
The cAMP/protein kinase A (PKA) signaling pathway regulates capsule size, mating, melanin formation, and virulence in C. neoformans (D'Souza et al., 2001; Alspaugh et al., 2002; Loftus et al., 2005; Hu et al., 2007; Kronstad et al., 2011a; 2011b; O'Meara et al., 2010; Choi et al., 2012). Disruption of the regulatory subunit of PKA (Pkr1) results in elevated PKA activity and an enlarged capsule phenotype, while disruption of the catabolic subunit of PKA (Pka1) causes reduced PKA activity and an acapsular phenotype (D'Souza et al., 2001). The cAMP/PKA pathway also activates Rim101 in C. neoformans (O'Meara et al., 2010). We deleted the genes VPS27, VPS23, VPS22, VPS20, SNF7, BRO1 and VPS4 in the pkr1 mutant, and examined the cells of the double mutants for capsule formation after growth on the defined LIM. Interestingly, cells of the vps23pkr1, vps22pkr1, vps20pkr1 and snf7pkr1 mutants displayed reduced capsule sizes compared with the pkr1 mutant (Figs. 5A, 5B, 5C), while the vps27pkr1, vps4pkr1 and bro1pkr1 double deletion mutants showed a similar capsule size to that of the pkr1 mutant (data not shown). Measurements of >50 cells of each strain revealed statistically significant differences in capsule sizes between the double mutants and the WT or pkr1 strains, and between the WT and the pkr1 mutant (Fig. 5C). These observations suggest that the ESCRT machinery functions downstream of, or in parallel with PKA to influence capsule size.
Figure 5. Defects in ESCRT machinery influence capsule formation in a pkr1 mutant lacking the regulatory subunit of PKA.
A. Capsule size was examined for the WT and pkr1 strains by growing cells in defined low iron medium at 30°C for 48h and staining with Indian ink. The pkr1 mutant is hyper-capsular compared with the WT strain.
B. Capsule formation by cells defective in ESCRT-I, -II and –III components in the pkr1 strain background. The top panel shows the capsule size for the double mutants upon India ink staining and the bottom panel shows the increase in capsule size detected upon overexpression of the N-terminal region of Rim101 from the RIM1011-628 allele.
C. Fifty cells of each strain were measured from the experiment in B to determine cell diameter and capsule radius. Each bar represents the average of the 50 measurements with standard deviations. The pEF1p designation indicates that the strains were transformed with the empty vector used for overexpression of the RIM1011-628 allele. The differences in capsule size for the pkr1 mutant (double asterisk) and the empty vector transformants (single asterisk) relative to all other strains are statistically significant by the Student t test (P<0.05).
We also tested the influence of Rim101 in the double mutants and found that introduction of the overexpression construct for the RIM1011-628 allele into the mutants (vps23pkr1, vps22pkr1, snf7pkr1 and vps20pkr1) restored the capsule formation almost to the WT level (Figs. 5A, 5B and 5C), but not to the size of the pkr1 mutant. The ESCRT machinery therefore contributes to capsule elaboration in part through an interaction with cAMP signaling and Rim101, and in part through a Rim101-independent connection with PKA. We also explored the influence of the RIM1011-628 allele in the acapsular pka1 mutant. We found that introducing the RIM1011-628 allele into this strain did not influence capsule formation (data not shown). This result also supports the hypothesis that PKA influences capsule formation and attachment via Rim101-dependent and independent mechanisms. It is possible that an ESCRT-PKA-Rim101 pathway influences attachment of capsular material to the cell wall, while an Rim101-independent pathway involving ESCRT and PKA influences other processes such as biosynthesis or trafficking of capsule polysaccharide. Additional work will be needed to examine the proposed Rim101-independent pathway and to rule out potential confounding issues such as a possible interaction between the Rim1011-628 and WT Rim101 proteins in these experiments that could potentially interfere with capsule elaboration.
The ESCRT machinery influences haem uptake by Rim101-dependent and independent mechanisms
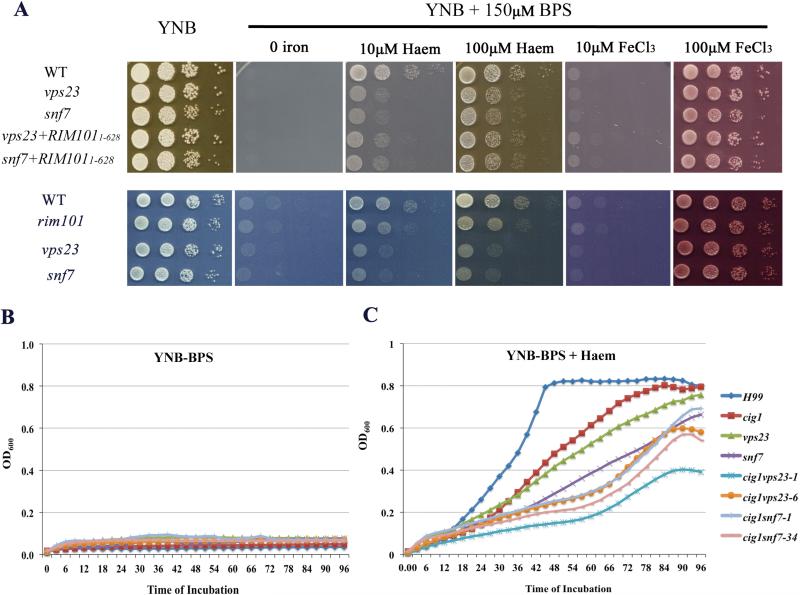
We showed previously that Rim101 is involved in iron acquisition from haem (Cadieux et al., 2013). The Rim101 pathway also regulates the expression of iron-responsive genes including CIG1, and deletion of CIG1 results in delayed growth on haem at neutral pH (Cadieux et al., 2013; O'Meara et al., 2010). Given these connections, we tested whether the involvement of the ESCRT machinery in haem uptake is controlled mainly via activation of Rim101. Specifically, we compared the growth of the WT strain, the vps23 and snf7 mutants, and the mutants carrying the RIM1011-628 allele on low iron medium with or without haem or FeCl3. As shown in figure 6A, the vps23 and snf7 mutants, and the mutants with the RIM1011-628 allele grew as well as the WT strain on YNB, YNB-LIM, or the low iron medium supplemented with either 10 μM or 100 μM FeCl3. On low-iron medium with the addition of either 10 μM or 100 μM haem, the vps23 and snf7 mutants with the RIM1011-628 allele showed only a subtle increase in growth compared with the original deletion mutants. That is, expression of the RIM1011-628 allele failed to restore growth to the WT level thus suggesting that the ESCRT machinery plays a role in heam acquisition beyond activation of Rim101. A comparison of the growth of the vps23, snf7 and rim101 mutants on haem is also shown in figure 6A.
Figure 6. ESCRT machinery influences iron use from haem via Rim101-dependent and independent mechanisms.
A. Expression of the N-terminal region of the RIM1011-628 allele from the native promoter at the RIM101 locus is unable to fully restore the growth defects of vps23 and snf7 mutants on low iron medium supplemented with haem. After iron starvation, ten-fold serial diluted cells of the indicated strains were spotted on the plates of low iron medium with or without the presence of haem or FeCl3. The plates were incubated at 30°C for 2 days prior to be photographed.
B and C. Iron-starved cells of the indicated strains were added to the liquid low iron medium (YNB plus 150 μM BPS) (B) or to the liquid low iron medium (YNB plus 150 μM BPS) with addition of 10 μM haem (C). Cells were grown at 30°C with agitation for 4 days and the OD600 was measured every 6 hours. The experiments were repeated three times with similar results.
We also deleted VPS23 and SNF7 in the cig1 mutant background and tested the resulting double mutants for growth on haem. Two independent double mutants for each gene were tested and cells were pre-starved for iron. As expected, none of the strains grew on low iron medium (YNB-LIM) and the cig1, vps23 and snf7 mutants showed impaired growth compared with the WT strain on low-iron medium supplemented with haem (Figs. 6B and 6C). However, the two independent double mutants for each combination (cig1vps23 and cig1snf7) showed additive growth defects on haem compared to the single mutants (Fig. 6C). These results suggest that the ESCRT functions can act independently of the Rim101/Cig1 pathway to influence haem uptake, perhaps by participating in endocytosis. Taken together, we conclude that the ESCRT machinery contribute to iron acquisition from haem via Rim101/Cig1-dependent and independent mechanisms.
The Rim101-independent mechanism does not appear to include a contribution to transcriptional regulation beyond the influence on Rim101 activation, based on an analysis of CIG1 transcription. That is, we compared the transcript levels for CIG1 in the WT strain and the ESCRT mutants in iron-replete and iron-limited conditions (Supplemental Fig. S6). We found that the transcript levels for CIG1 were still responsive to iron deprivation in the ESCRT-I, II and III mutants, albeit to a reduced level compared with the WT strain (Supplemental Fig. S6). Therefore, the ESCRT functions were not essential for at least part of the transcriptional response to iron deprivation.
The ESCRT machinery is implicated in cryptococcal survival in macrophages and virulence
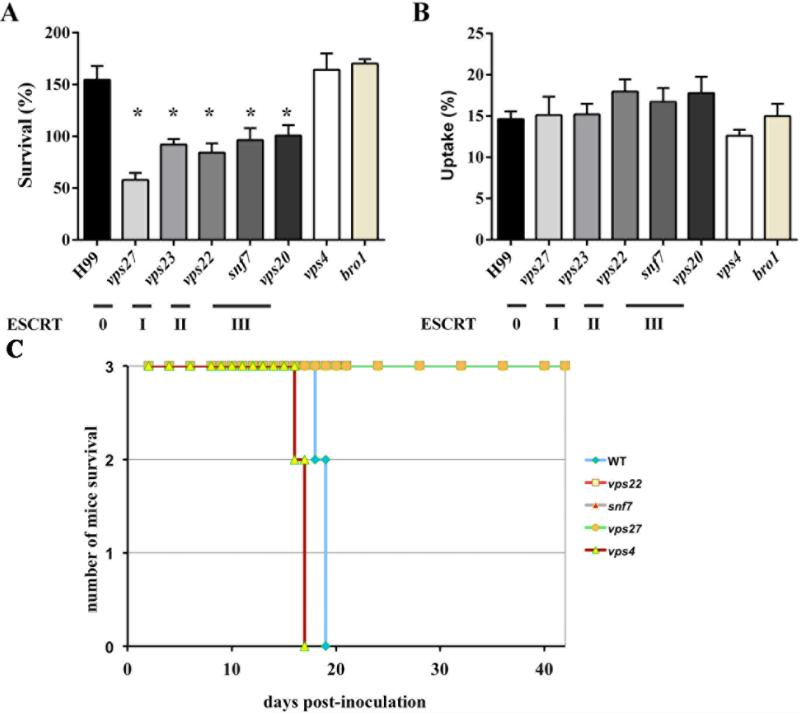
We also examined the ability of the ESCRT mutants to survive in the presence of the J774A.1 macrophage-like cell line. The macrophage cells were incubated with opsonised cells of the WT strain and the ESCRT mutants at a multiplicity of infection (MOI) of 1:1 to examine uptake and intracellular survival. Survival was expressed as the ratio of CFU ml−1 obtained from macrophage lysis after 24h incubation compared to an initial incubation of 2h. The vps27, vps23, vps22, snf7 and vps20 mutants all had impaired intracellular survival compared to the WT strain (Fig. 7A). The CFUs obtained after 24h for these mutants were ~50% compared to their initial count at 2h and 160% for the WT strain (Fig. 7A). The vps4 and bro1 mutants retained the ability to survive inside macrophages to a level comparable to the WT strain (Fig. 7A). All of the mutants, including those displaying a difference in survival, were taken up at a similar rate to the WT (Fig. 7B). Remarkably, the vps27 (ESCRT-0) showed the most significant reduction in survival when phagocytosed by the J774A.1 cells.
Figure 7. Defects in ESCRT components influence survival in macrophages and virulence in mice.
A-B. Intracellular survival and uptake of the WT strain and the ESCRT mutants inside J774A.1 macrophage-like cell line.
A. Intracellular survival of the wild type and the ESCRT mutants in J774A.1 macrophage-like cell line (* P < 0.01 relative to survival of the WT strain). The WT strain and the mutants were incubated for 1 h and 24 h in DMEM containing macrophages at a MOI of 1:1 at 37°C with 5% CO2. Experiments were performed at least three times in triplicate and with two different strains carrying the same mutation; representative data for one mutant is shown. Values are expressed as the mean ± standard error of the mean.
B. Phagocytic uptake of the WT strain and the ESCRT mutants after 1 h of interaction with the macrophage cell line at a MOI of 1:1. No statistically significant differences in uptake were noted.
C. The WT strains and the vps22, vps27, snf7 and vps4 mutants were tested in a virulence assay with BALB/c mice. Three mice were inoculated intranasally with each of the strains at 106 cells in 50 μl PBS. The survival of the mice was monitored over time. The vps4 was as virulent as the WT strain and the other three strains showed complete attenuation of virulence.
We previously demonstrated that deletion of the gene for the ESCRT-I component Vps23 caused avirulence in a mouse inhalation model (Hu et al., 2013). As described above, mutants lacking components of the ESCRT-II and –III complexes displayed similar phenotypes to the vps23 mutant including reduced production of capsule and melanin, reduced growth on haem, and reduced survival in macrophages. We therefore predicted that mutants representative of the ESCRT-II and III complexes (e.g., vps22 and snf7) would be avirulent. We confirmed this prediction with small-scale virulence assays in mice (Fig. 7C) and additionally found that the vps27 (ESCRT-O) mutant, but not the vps4 mutant, was avirulent. We also note that a recent report by Godinho et al. (2014) described an avirulent phenotype for a snf7 mutant of C. neoformans. Overall, we conclude that the ESCRT-0, -I, II, and –III proteins are required for virulence in mice as well as survival in macrophages.
Discussion
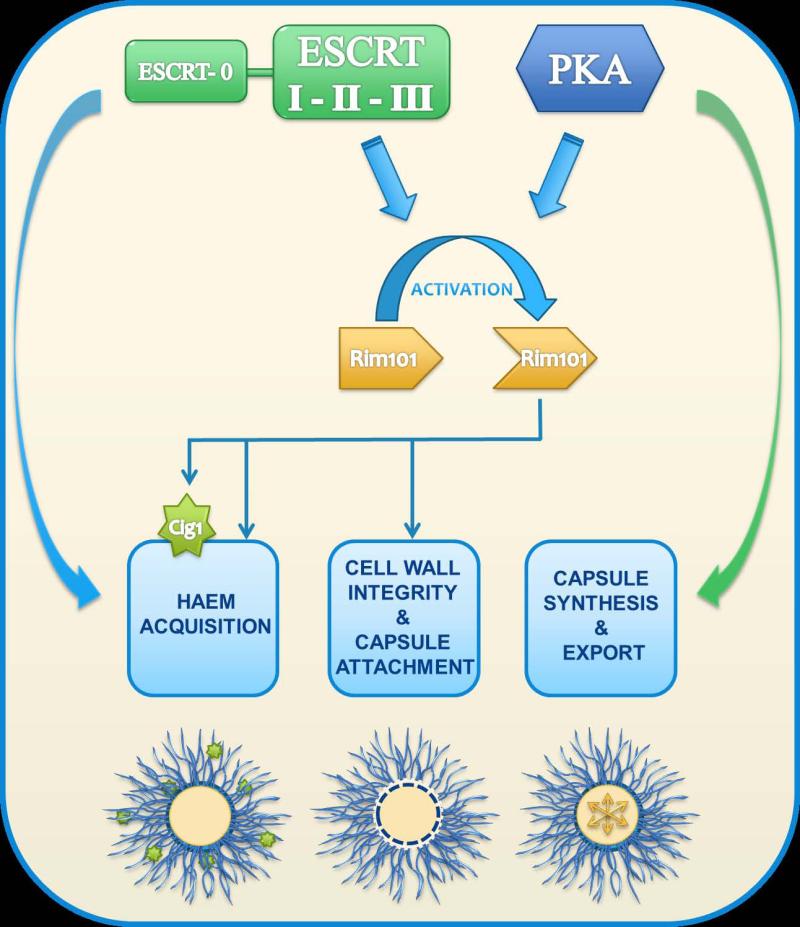
In this study, we extended our analysis of the ESCRT machinery and found that mutants lacking representative ESCRT-II and –III components share the haem and capsule phenotypes previously discovered for the vps23 mutant (Hu et al., 2013). The C. neoformans mutants also shared conserved phenotypes with comparable ESCRT mutants in other fungi such as S. cerevisiae, C. albicans and Aspergillus nidulans (Xu et al., 2004; Penalva et al., 2008). These included an involvement in endosomal trafficking, as demonstrated by the accumulation of E-like bodies surrounding the vacuoles of the mutants. Furthermore, the C. neoformans mutants in the core ESCRT machinery displayed Rim101-dependent phenotypes and we confirmed the physical interaction between the ESCRT-III protein Snf7 and Rim20, a regulatory protein in the Rim101 pathway. This interaction contributes to the proteolytic cleavage and activation of Rim101 in other fungi, and Rim101 processing has also been demonstrated in C. neoformans (O'Meara et al., 2010). The conservation of this process in C. neoformans is consistent with the shared phenotypes of rim101 and ESCRT mutants in capsule attachment to the cell wall, the regulation of iron-responsive functions such as the candidate haem-binding protein Cig1, and growth on haem (O'Meara et al., 2010, Cadieux et al., 2013). Activation of Rim101 is also dependent on PKA in C. neoformans and, in this context; we found that the core ESCRT machinery act downstream of or in a parallel pathway with PKA to regulate capsule size. Together these observations support a model in which PKA and the ESCRT machinery act together to activate Rim101 in response to signals from the environment (e.g., pH) (Fig. 8). Activated Rim101 then regulates downstream functions involved in cell wall formation to support the attachment of capsule polysaccharide and to regulate the expression of proteins such as Cig1 that influence haem uptake. We used an N-terminal portion of Rim101 to further determine whether the ESCRT machinery and PKA played roles beyond the activation of Rim101. This approach provided further resolution for the model by revealing Rim101-independent contributions of the ESCRT machinery to haem uptake and PKA contributions to capsule formation.
Figure 8. Proposed model for the influence of the ESCRT machinery on capsule formation and haem uptake.
In response to environmental or host signals, the core ESCRT machinery (ESCRT-I, II and III) participates with PKA to activate Rim101 and influence capsule attachment via regulation of cell wall composition. The bulk of the ESCRT influence on capsule formation occurs through activation of Rim101 given the shared defects in attachment. PKA may make a contribution to capsule formation in addition to its influence on Rim101 because expression of the N-terminal portion of Rim101 in double mutants defective for both the regulatory subunit of PKA and ESCRT functions (e.g., vps23 pkr1) did not restore capsule size to the level of the pkr1 mutant (Fig. 5). Additionally, the cAMP/PKA pathway is known to influence the transcription of genes for capsule synthesis as well as components of the secretory pathway (Pukkila-Worley et al., 2005; Hu et al., 2007). The ESCRT machinery contributes to iron acquisition from haem via Rim101 and Cig1-dependent and independent mechanisms. ESCRT mutants have a growth defect on haem and expression of the N-terminal portion of Rim101 in the mutants only slightly improved growth. The remaining growth defect may reflect a requirement for ESCRT participation in endocytosis of haem, and the minor improvement with activation of Rim101 may in part reflect the expression of Cig1 for haem uptake. The involvement of PKA is more complex because pka1 and pkr1 mutants do not show growth defects on haem (G. Hu, unpublished data).
Iron acquisition is critical for pathogens such as C. neoformans to overcome nutritional immunity in vertebrate hosts (Cassat and Skaar, 2013). However, the molecular basis for the uptake and use of haem, the most abundant iron source in mammals, is only just now being explored in fungal pathogens (Jung and Kronstad, 2008; Kronstad et al., 2013; Kuznets et al., 2014). Several fungal pathogens such as C. albicans and Histoplasma capsulatum can use haem and hemoglobin as an iron source (Foster, 2002; Santos et al., 2003). Mutations in the ESCRT machinery in C. albicans cause growth defects on hemoglobin as the sole iron source indicating that the involvement of ESCRT machinery in haem uptake and utilization is a conserved function in the pathogenic fungi (Weissman and Kornitzer, 2004; Weissman et al., 2008). However, there are differences between fungi. For example, C. albicans mutants lacking ESCRT-II components (Vps22 of Vps25) or ESCRT-III components (Snf7 or Vps20) displayed impaired growth on haem or haemoglobin, but none of the ESCRT-I mutants showed significant growth defects on haem (Weissman et al., 2008). In contrast, we found that mutations in C. neoformans ESCRT-I, -II or –III components all impaired growth on haem, and these proteins likely play a role in uptake because the mutants had reduced susceptibility to noniron MPs that require a haem uptake system for toxicity.
The connections between ESCRT functions and iron acquisition occur at multiple levels including transcriptional control via Rim101 and trafficking of iron transport proteins. With regard to transcriptional control, we found that none of the mutations in ESCRT machinery components fully blocked the increase in CIG1 transcript level upon iron deprivation, although a reduced level of induction was observed perhaps reflecting the contribution to Rim101 activation. Our previous work revealed that CIG1 transcript levels are greatly elevated upon iron limitation and that the transcript is the most abundant in cells grown in this condition (Lianet al., 2005). As mentioned, CIG1 is highly regulated by Rim101 under certain conditions with a ~ 470-fold reduction in the rim101 mutant (O'Meara et al., 2010). Consistent with this regulation, we have shown that deletion of either RIM101 or CIG1 leads to impaired growth on haem (Cadieux et al., 2013). However, expression of Cig1 from a Rim101-independent promoter in a rim101 mutant revealed that Rim101 makes additional contributions to growth on haem beyond activating expression of CIG1 (Cadieux et al., 2013; Fig. 8). In addition, the defect in growth on haem for the rim101 or cig1 mutants is less severe than for the ESCRT mutants (O'Meara et al., 2010; Cadieux et al., 2013). We therefore hypothesize that the contribution of the ESCRT machinery for haem occurs only in part via activation of Rim101 leading to expression of Cig1 (Fig. 8). This hypothesis is also supported by the fact that expression of the N-terminal region of Rim101 in the snf7 and vps23 mutants had little impact on the growth of the mutants on haem. The additive growth defects of the cig1vps23 and cig1snf7 double mutants on haem, relative to the single mutants, also support a role for the ESCRT machinery that is independent of Rim101 and Cig1. This hypothesis is consistent with the weak restoration of growth defects of vps23 and snf7 upon expression of the N-terminal region of Rim101. Overall, this work highlights the interactions between the ESCRT machinery and Rim101 in the use of haem via Cig1 dependent and independent mechanisms.
Additional roles for the ESCRT machinery in iron use from haem likely involve endocytosis and intracellular trafficking. The ESCRT machinery generally targets monoubiquitinated membrane proteins for internalization and trafficking to the vacuole (Hurley and Emr, 2006). Although Cig1 may be targeted for endocytosis, it contributes to haem uptake only at neutral pH (Cadieux et al., 2013). Therefore additional receptor(s) for haem may be present in C. neoformans and these could be monoubiquinated and serve as targets for the ESCRT machinery. The ESCRT-II and –III components in C. albicans play a role in the endocytic pathway for iron acquisition from haemoglobin, and the GPI-anchor proteins Rbt5 and Pga7 function as haemoglobin/haem receptors (Weissman et al., 2008; Kuznets et al., 2014). Endocytosis may also be involved in the use of other iron sources like siderophores, e.g., as found for the Arn1-dependent ferrichrome uptake system in fungi (Kim et al., 2002, Hu et al., 2002). However, our ESCRT mutants did not show defects in iron acquisition from ferrichrome suggesting that endocytosis is not required for iron acquisition from this siderophore.
We also found that the core components of ESCRT-I, -II, and III are required for the elaboration of capsule, a major virulence factor in C. neoformans. The reduced capsule size in the ESCRT-I, -II and –III mutants is due to poor attachment of polysaccharide to the cell wall because the mutants shed polysaccharide and this material was still competent for attachment to an acapsular strain. Altered cell wall composition in the mutants is consistent with their susceptibility to agents like congo red and caffeine that challenge cell wall integrity. O'Meara et al. (2010) previously established that the reduced capsule size of the rim101 mutant is due to shedding rather than decreased synthesis. We were able to restore capsule attachment and reduce shedding by expression of the N-terminal portion of Rim101 in the rim101 and the ESCRT-I, -II and –III mutants. Therefore, we propose that the bulk of the ESCRT influence on capsule occurs through activation of Rim101. However, a role for ESCRT proteins in the secretion of capsular material to influence capsule size is also possible, as suggested by Godinho et al. (2014).
The cAMP-PKA pathway plays an important role in capsule formation and virulence in C. neoformans (D'Souza et al., 2001; Hicks et al., 2004; Pukkila-Worley et al., 2005; Hu et al., 2007; Choi et al., 2012). O'Meara et al. (2010) established that PKA regulates Rim101 through an influence on localization and activation. In our study, we observed that the deletion of ESCRT-I, -II and –III components in the pkr1 mutant resulted in a significant reduction in capsule size. This result suggests that the ESCRT machinery act downstream of PKA activity to influence capsule, perhaps via a link between Rim101 phosphorylation by PKA and activation by ESCRT-mediated cleavage. We also found that overexpression of the N-terminal portion of Rim101 restored capsule formation in the double mutants lacking Pkr1 and either Vps23 or Snf7, although only to a size like WT and not to the level of the pkr1 mutant. Notably, our N-terminal portion of Rim101 (aa 1-628) lacked the C-terminal D/E rich region including the putative PKA phosphorylation site (aa 773). This version might therefore bypass both PKA phosphorylation and the cleavage needed for Rim101 activation. The incomplete restoration of capsule size to the level of the pkr1 mutant suggests that PKA makes an additional contribution beyond the involvement of the ESCRT machinery and Rim101, as indicated in Fig. 8. This idea is also supported by the finding that expression of the N-terminal portion of Rim101 does not restore capsule formation in a pka1 mutant (G. Hu, unpublished results). We speculate that PKA may influence additional steps in capsule formation such as polysaccharide synthesis and trafficking to the cell surface as indicated in other studies of the cAMP/PKA pathway in C. neoformans (Pukkila-Worley et al., 2005; Hu et al., 2007).
Interestingly, the vps27 (ESCRT-0) mutant had unique phenotypes in C. neoformans. Deletion of VPS27 did cause defective endosome trafficking as revealed by the abnormal E-like bodies around the vacuoles as well as increased sensitivity to NaCl, LiCl and alkaline pH. However the growth defect in haem was minor compared with the ESCRT-I, II, III mutants. Additionally, cells of the vps27 mutant had a WT capsule size, although they were larger than the WT cells or the cells of the ESCRT-I, II and III mutants. Importantly, the vps27 mutant was attenuated for virulence in mice and showed reduced survival in macrophages. These phenotypes suggest other functions for Vps27 in C. neoformans that may reflect the ability of Vps27 to bind polyubiquitinated proteins (Ren and Hurley, 2010). By contrast, the bro1 and vps4 mutants behaved like the WT strain in terms of growth on haem, capsule formation, sensitivity to the salt stress and alkaline pH, virulence in mice, and survival in macrophages. Vps4 catalyzes disassembly of the ESCRT-III complex and is not required for activating the Rim101 pathway. Rather, it was observed in S. cerevisiae that disruption of VPS4 leads to the constitutive activation of Rim101 (Hayashi et al., 2005). Bro1 acts as an accessory protein and also does not influence activation of Rim101 (Boysen et al., 2010). Therefore, the different phenotypes of the vps4 and bro1 mutants are consistent with distinct functions relative to the other ESCRT proteins.
We also found that the core ESCRT components play a role in the virulence in C. neoformans. These results confirmed previous virulence assays that revealed that vps23 and snf7 mutants are avirulent in a mouse inhalation model (Hu et al., 2013; Godinho et al., 2014). Capsule formation and iron acquisition are important for cryptococcal pathogenesis and the defects in these traits may account in part for the attenuated virulence of the ESCRT mutants (Kronstad et al., 2012; O'Meara and Alspaugh, 2012; Kronstad et al., 2013). Interestingly, the rim101 mutant exhibited increased virulence in AJc/r mice or similar virulence in C57BL/6 mice compared with the WT strain (O'Meara et al., 2010; O'Meara et al., 2013). Although the ESCRT machinery functions upstream of Rim101 pathway to influence capsule elaboration, salt stress and iron utilization (partially), it also plays a broader role in iron acquisition from haem that is independent of the Rim101 pathway. Moreover, the ESCRT machinery acts in targeting membrane proteins (e.g., cell surface receptors and transporters) to the vacuole for degradation, and it is required for other cellular processes independent of the Rim101 pathway. These other functions likely also contribute to the ability of the fungus to cause disease. Similarly, defects in ESCRT components in C. albicans also cause reduced virulence in animal models, and ESCRT mutants were less virulent than a rim101 mutant indicating an influence beyond the Rim101 pathway (Cornet et al., 2005; Davis, 2009; Wolf et al., 2010).
Experimental Procedures
Strains, plasmids, and media
The serotype A strain H99 (C. neoformans var. grubii) was used for all experiments and was maintained on YPD medium (1% yeast extract, 2% peptone, 2% dextrose, 2% agar). Yeast nitrogen base-low-iron medium (YNB-LIM) (composed of YNB plus 150 μM bathophenanthroline disulfonate (BPS) at pH 7.2) (Jung et al., 2010) and defined low-iron medium (LIM) were prepared as described previously (Lian et al., 2005; Griffiths et al., 2012). These media were used as iron-limiting media and supplemented as indicated, for phenotypic characterization. YNB-LIM was prepared in iron-chelated water using Chelex-100 (Invitrogen), adjusted to pH 7.0 with 3-morpholinopropanesulfonic acid (MOPS), and iron limitation was achieved with 150 μM BPS. Defined LIM to induce the capsule formation was composed of 0.5% glucose, 38 mM L-asparagine, 2.3 mM K2HPO4, 1.7 mM CaCl2·2H2O, 0.3 mM MgSO4·7H2O, 20 mM HEPES, 22 mM NaHCO3 plus 1 ml l−1 1000X salt solution (0.005 g l−1 CuSO4·5H2O, 2 g l−1 ZnSO4·7H2O, 0.01 g l−1 MnCl3·4H2O, 0.46 g l−1 sodium molybdate, 0.057 g l−1 boric acid in 1 l of iron-chelated H2O). The solution was iron chelated (BIORAD chelex-100), the pH was adjusted to 7.4, and filter sterilized. Finally, 0.4 mg l−1 thiamine was added. All chemicals were obtained from Sigma-Aldrich unless indicated otherwise. Haem was added to cultures as hemin (Sigma 51280, FW651.94) and is referred as haem throughout the manuscript. Staining of cells with lipophilic dyes FM4-64 and MDC64 was performed to observe internalized vesicle trafficking. The cells were stained with the dyes on ice for 15 min (at a final concentration of 10 μM for FM4-64 and 5 μM for MDC64) and washed to remove the unbound dye. The cells were incubated for an additional 30 min at 30°C and then visualized using an Axioplan 2 imaging microscope (Zeiss).
Identification of ESCRT genes and construction of deletion mutants
The components of ESCRT complex in C. neoformans were identified by a BLASTp search of the H99 genome database (www.broadinstitute.org) using the proteins of the ESCRT complexes from S. cerevisiae, as listed in Table 1. All deletion mutants were constructed by homologous recombination using gene specific knockout cassettes, which were amplified by three-step overlapping PCR (Hu et al., 2008) with the primers listed in Supplemental Table S1. The resistance markers for nourseothricin (NAT), neomycin (NEO), and hygromycin (HYG), were amplified by PCR using the primers 2 and 5 and the plasmids pCH233, pJAF1, and pJAF15, respectively, as the templates. In general, the gene-specific knockout primers 1 and 2 and 4 and 6 were used to amplify the flanking sequences of their respective genes; and the primers 1 and 6 were used to amplify the gene-specific deletion construct containing the resistance marker. All constructs for deletions were introduced into the H99 wild-type (WT) strain, or a pkr1 mutant of the H99 strain, by biolistic transformation, as described previously (Davidson et al., 2000).
Growth assays
To assess growth on solid media, 10-fold serial dilutions of cells were spotted on agar plates with or without supplemented iron sources. Plates were incubated at 30°C for 2 days (or as indicated) before being photographed. Growth of the strains was also assessed in 96-well microplates using the Infinite M200 PRO plate reader (Tecan, Austria). For this assay, iron-starved cells were inoculated in 200 μl of YNB-LIM with or without supplemented iron sources for 5 days at 30°C (final inoculum of 104 cells). Growth was monitored by measuring the optical density at 600 nm, and the resulting data were analyzed using the Magellan software (Tecan, Austria).
Toxicity of gallium protoporphyrin and manganese protoporphyrin
To test for GaPPIX and MnPPIX toxicity, cells grown overnight in YPD medium were washed twice in sterile, low-iron water (treated with Chelex-100 resin). Tenfold serial dilutionsof the cells were prepared in low-iron water. Defined LIM agar containing 10 μM haem was spread with 200 μl of 10 μM GaPPIX (Frontier Scientific), 100 μM MnPPIX (Frontier Scientific), or 10 μM GaCl3 (Sigma-Aldrich) immediately prior to the spotting of 5μl volumes of dilutions of 1 ×106 to 1 ×102 cells ml−1. The plates were incubated for 2 days at 30°C before being photographed.
Capsule formation and melanin production
Capsule formation was examined by differential interference contrast microscopy after incubation for 24 h at 30°C in defined LIM and staining with India ink. Capsule shedding from cells was examined with an immunoblot assay and anti-capsule mAb 18b7, as described by Yoneda and Doering (2008). The capsule polysaccharide transfer assay was conducted with a modified version of published protocols (Reese and Doering, 2003; O'Meara et al., 2010; Fang et al., 2012). Briefly, cells were grown in defined LIM at 30°C for 3 days, and the supernatants were collected from 1.5ml of cells of each strain. For capsule transfer assays, acapsular cells of the cap59 mutant (in strain KN99a) were grown in YPD and used as acceptors of polysaccharide from other strains. The cap59 cells were washed with PBS (pH 7.4), and re-suspended in 2 ml of PBS. A 50 μl sample of the cap59 cells was then incubated with 1 ml of supernatant from each strain at 30°C for 2h (with shaking). The cells were then washed with PBS and incubated with mAb 18b7 (0.1~0.2 mg/ml, 20X dilution of the antibody; a generous gift from Dr. Arturo Casadevall) for 2 h. The cells were washed again with PBS and further incubated with secondary antibody (Alexa Fluor anti-mouse IgG, Invitrogen, diluted 50~100X times from the stock of 2 mg l−1) for 1h. The cells were then examined for fluorescence with an Axioplan 2 imaging microscope (Zeiss). Melanin production was examined on L-3,4-dihydroxyphenylalanine (L-DOPA) plates containing 0.1% glucose.
Yeast two-hybrid assay
The assays were performed using the ProQuest™ Two-Hybrid System and methods from Invitrogen. Briefly, RIM20 and SNF7 were PCR amplified from C. neoformans cDNA and cloned into pDEST32 (bait) and pDEST22 (prey) vectors, respectively. Growth of MaV203 yeast expressing both bait and prey vectors was tested on SC medium lacking leucine and tryptophan to select for each vector, and histidine and uracil to test for an interaction. Empty pDEST32 and pDEST22 vectors were used as negative controls. Quantification of β-galactosidase activity was performed using cells collected from overnight cultures.
Overexpression of a truncated allele of RIM101 and replacement of the native RIM101 allele with the truncated allele
We identified an N-terminal fragment of Rim101 (allele designation RIM1011-628) as a candidate for the activated form of the protein by evaluation of information on Rim101 cleavage in C. neoformans and other fungi (Lambert et al., 1997; O'Meara et al., 2010). To prepare an overexpression construct of RIM1011-628, PCR of genomic DNA was used to amplify part of the RIM101 gene encoding an N-terminal polypeptide of 68 kD using primers N-Rim101-P1F /P1Ra or N-Rim101-P1F/P1Rb, or N-Rim101-P1F/P1Rc, respectively. Another fragment of 442 bp was amplified for the terminator region of the RIM101 gene using primer pairs N-Rim101-P2Fa/N-Rim101P2R, N-Rim101-P2Fb/N-Rim101P2R or N-Rim101-P2Fc/N-Rim101P2R, respectively. The stop codon TAG was added to the end of each truncated fragment (at the beginning of the RIM101 terminator region). An overlap PCR reaction was subsequently performed, and the products were digested with XbaI and cloned into SpeI cut pEF1pro-Vps23-ter (Hu et al., 2013). Plasmid DNA with the correct orientation was linearized with BglII before being biolistically transformed into the WT and mutant strains (H99, vps22, vps23, snf7, vps20, pka1, rim101 and pkr1), respectively. The transformants were screened by colony-PCR after growth on the hygromycin B plates to identify the strains containing the RIM101 gene encoding the N-terminal region of the protein, as expressed from the constitutive promoter of elongation factor I (Hu et al., 2013).
An overlap PCR strategy was used to generate the strains expressing the 65 kD N-terminal region of Rim101 from the native promoter. Briefly, the construct to express the N-terminal, 68 kD, portion of Rim101 was prepared by amplification of a DNA fragment of 926 bp from H99 genomic DNA using primers Rim101-NN-P1Fb and Rim101-NN68K-P1R. A fragment of 2,717 bp including the GAL7-terminator and the hygromycin B resistance marker was amplified from plasmid pGH023 using the primers Rim101-NN68K-P2F and Rim101-NN68K-P2R. The plasmid pGH023 containing the GAL7-terminator was made by cloning a HindIII/SpeI digested PCR product amplified using genomic DNA as the template into the HindIII/SpeI cut pJAF15; and a fragment of 485 bp downstream of the RIM101 gene from genomic DNA using primers Rim101-NN-P4F and Rim101-NN-P4R. The overlapping PCR was performed to amplify the final construct of 4128 bp using primers Rim101-NN-P1Fb and Rim101-NN-P4R. The construct was introduced into the WT, vps23-neo and snf7-neo strains by biolistic transformation. Replacement of the native RIM101 allele with the truncated allele was confirmed by colony PCR on transformants from medium containing 200 μg ml−1 of hygromycin B and 200 μg ml−1 of G418.
Real time-PCR
Real-time PCR analysis was conducted as previously described (Hu et al., 2007) using primers designed with PrimerExpress (Applied Biosystems, http://www.appliedbiosystems.com). Briefly, total RNA from frozen cells was extracted the using RNeasy mini kit (Qiagen), DNA was removed by treatment with Turbo Dnase (Ambion) for 30 min at 25°C, and cDNA was synthesized using a mixture of anchored oligo dT random hexamers (3:1), and Superscript transcriptase II (Invitrogen Canada). The resulting cDNA was used for real-time PCR with Power SYBR Green PCR mix (Applied Biosystems) according to the manufacturer's recommendations. An Applied Biosystems 7500 Fast Real-Time PCR System was used to detect and quantify the PCR products using the following conditions: incubation at 95°C for 10 min followed by 40 cycles of 95°C for 15 sec, and 60°C for 1 min. The cDNA of the 18S RNA gene was used to normalize the data. Dissociation analysis on all PCR reactions confirmed the amplification of a single product for each primer pair and the lack of primer dimerization (Applied Biosystems). The primers for each gene are listed in Supplemental Table S1. Relative gene expression was quantified using SDS software 1.3.1 (Applied Biosystems) and the 2−ΔΔCt method (Livak and Schmittgen, 2001).
Macrophage survival assay and the virulence assay in a mouse inhalation model
The survival of WT and mutant strains during incubation with macrophages was assessed as previously described (Hu and Kronstad, 2010; Griffiths et al., 2012; Kretschmer et al., 2014). Briefly, the murine macrophage-like cell line J774A.1 was maintained at 37°C in 5% CO2 in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% heat-inactivated fetal calf serum (FBS), 100 μg ml−1 penicillin-streptomycin, and 4 mM L-glutamine (Invitrogen). The cell line was used between passages 5 and 10 for the assays. Macrophages were stimulated 2h prior infection with 150 ng mL−1 phorbol myristate acetate (PMA). Cells of the WT and mutant strains were opsonized with monoclonal antibody 18b7 against capsule (1 μg ml−1), and coincubated at a multiplicity of infection (MOI) of 1:1. Stimulated macrophages were inoculated with 2×105 of opsonized fungal cells and washed 3 times with PBS after 2 h of inoculation to remove unattached, extracellular fungal cells. After 24 h of incubation, sterile distilled H2O was applied to each well to lyse the macrophages (confirmed microscopically). Fungal growth and survival was measured by plating on YPD and determining CFUs. The assay was performed in triplicate wells for each strain, and the experiment was repeated three times with consistent results. To measure uptake rate of cryptococcal cells by macrophages, opsonized cells were incubated with stimulated macrophages for 2h at 37°C with 5% CO2. Cells were then washed 3 times with PBS, fixed with 4% paraformaldehyde, washed two more times with PBS and mounted on microscopic slides. Macrophages containing fungal cells were quantified by DIC microscopy (100X magnification). A minimum of 500 cells were count per replicate. Statistical significance of intracellular survival and uptake rate was determined by unpaired t-tests. * P < 0.01
Virulence assays with a mouse inhalation model were conducted as previously described (Hu et al., 2013). Briefly, C. neoformans strains were grown in 5 ml of YPD medium at 30°C overnight, washed twice with PBS (Invitrogen), and re-suspended in PBS. Three BALB/c mice for each strain were intranasally inoculated with a suspension of 106 cells in 50 μl. The health status of the mice was monitored daily post-inoculation. Mice reaching the humane endpoint were euthanized by CO2 anoxia. The protocol for the virulence assay (protocol A13-0093) was approved by the University of British Columbia Committee on Animal Care.
Supplementary Material
Supplemental Figure S1. Defective endocytosis and increased susceptibility to trafficking inhibitors for the ESCRT mutants. A. The WT and ESCRT mutant strains were grown in YPD medium at 30°C overnight, harvested, washed in PBS (pH 7.2), and stained with MDY-64 on ice for 15 min and washed to remove the unbound dye. The cells were then incubated for an additional 30 min at 30°C before viewing. The DIC (differential interference contrast) images are shown on the left and the fluorescence resulting from MDY64 staining is shown on the right. The results were also confirmed with FM4-64 staining (data not shown). B. Ten-fold serial dilutions of the indicated strains were spotted onto YNB alone or YNB supplemented with either 20 μg ml−1 or 40 μg ml−1 brefeldin A (BFA). The plates were incubated for 3 days at 30°C.
Supplemental Figure S2. Requirement of ESCRT components for the growth in liquid low iron medium supplemented with haem. Cells were inoculated into the liquid YNB + 150 μM BPS with and without ferric chloride or haem (10μM and 100μM), and then incubated at 30°C for 5 days. Optical density at 600 nm (OD600) were measured every 4 h using a NanoQuant Infinite M200 Pro TECAN system. Experiments were performed in triplicated. Each point stands for the expression of the mean of three measurements ± standard deviation (SD).
Supplementary Figure S3. Comparison of haem rescue of fluconazole susceptibility for mutants defective in ESCRT-I or high affinity iron uptake functions. Suppression of the fluconazole susceptibility of the vps23, cfo1, vps23 cfo1 and rim101 mutants was tested by addition of exogenous haem at 10 μM to YNB plus BPS medium containing 10 μg ml−1 fluconazole. For all assays, the strains were grown in low-iron medium and ten-fold serial dilutions of were spotted on agar plates, with subsequent incubation for 2-3 days at 30°C.
Supplementary Figure S4. Susceptibility of ESCRT mutants to agents that challenge cell wall integrity. Ten-fold serial dilutions of cells of the indicated strains were spotted onto YPD medium containing either 0.5 mg ml−1 caffeine, 0.5 mg ml−1 congo red, or 0.05% sodium dodecylsulfate (SDS). The plates were incubated for 3 days at 30°C.
Supplementary Figure S5. Reduced melanisation in ESCRT mutants. Ten-fold serial dilutions of cells of the indicated strains were spotted onto solid L-DOPA plates. The plates were incubated for 2 days at 30°C or 37°C prior to being photographed. Note, the ESCRT-III mutant, snf7, exhibits weak growth defect on YPD at 37°C, and has the most significant defect in melanisation. The vps4 and bro1 mutants produced a similar level of melanisation to the WT strain.
Supplemental Figure S6. Comparison of transcript levels for iron acquisition and haem uptake functions in response to iron deprivation. RNA levels for CIG1 were measured by quantitative RT-PCR analysis and compared for cells of the WT strain and the ESCRT mutants grown in low iron medium (YNB + BPS) versus iron-replete medium (YNB). The assays were performed twice in triplicate, and relative fold change values are reported as the mean ± SEM. The dashed lines indicate a two-fold change in transcript levels.
Acknowledgements
This work was supported by grants from the Canadian Institutes of Health Research (JWK), the National Institutes of Health (RO1AI053721, JWK) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT, and Future Planning NRF-2013R1A1A1A05007037 (WHJ). The authors thank Dr. Arturo Casadevall for the 18b7 antibody. JWK is Burroughs Wellcome Fund Scholar in Molecular Pathogenic Mycology.
References
- Alspaugh JA, Pukkila-Worley R, Harashima T, Cavallo LM, Funnell D, Cox GM, et al. Adenylyl cyclase functions downstream of the Galpha protein Gpa1 and controls mating and pathogenicity of Cryptococcus neoformans. Eukaryot Cell. 2002;1:75–84. doi: 10.1128/EC.1.1.75-84.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchin-Roland S, Da Costa G, Gaillardin C. ESCRT-I components of the endocytic machinery are required for Rim101-dependent ambient pH regulation in the yeast Yarrowia lipolytica. Microbiology. 2005;151:3627–3637. doi: 10.1099/mic.0.28196-0. [DOI] [PubMed] [Google Scholar]
- Bowers K, Lottridge J, Helliwell SB, Goldthwaite LM, Luzio JP, Stevens TH. Protein-protein interactions of ESCRT complexes in the yeast Saccharomyces cerevisiae. Traffic. 2004;5:194–210. doi: 10.1111/j.1600-0854.2004.00169.x. [DOI] [PubMed] [Google Scholar]
- Boysen JH, Mitchell AP. Control of Bro1-domain protein Rim20 localization by external pH, ESCRT machinery, and the Saccharomyces cerevisiae Rim101 pathway. Mol Biol Cell. 2006;17:1344–1353. doi: 10.1091/mbc.E05-10-0949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen JH, Subramanian S, Mitchell AP. Intervention of Bro1 in pH-responsive Rim20 localization in Saccharomyces cerevisiae. Eukaryot Cell. 2010;9:532–538. doi: 10.1128/EC.00027-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassat JE, Skaar EP. Iron in infection and immunity. Cell Host Microbe. 2013;13:509–519. doi: 10.1016/j.chom.2013.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadieux B, Lian T, Hu G, Wang J, Biondo C, Teti G, et al. The mannoprotein Cig1 supports iron acquisition from heme and virulence in the pathogenic fungus Cryptococcus neoformans. J Infect Dis. 2013;207:1339–1347. doi: 10.1093/infdis/jit029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Vogl AW, Kronstad JW. Regulated expression of cyclic AMP-dependent protein kinase A reveals an influence on cell size and the secretion of virulence factors in Cryptococcus neoformans. Mol Microbiol. 2012;85:700–715. doi: 10.1111/j.1365-2958.2012.08134.x. [DOI] [PubMed] [Google Scholar]
- Chun CD, Madhani HD. Ctr2 links copper homeostasis to polysaccharide capsule formation and phagocytosis inhibition in the human fungal pathogen Cryptococcus neoformans. PLoS One. 2010;5 doi: 10.1371/journal.pone.0012503. pii: e12503. doi: 10.1371/journal.pone.0012503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornet M, Bidard F, Schwarz P, Da Costa G, Blanchin-Roland S, Dromer F, Gaillardin C. Deletions of endocytic components VPS28 and VPS32 affect growth at alkaline pH and virulence through both RIM101-dependent and RIM101-independent pathways in Candida albicans. Infect Immun. 2005;73:7977–7987. doi: 10.1128/IAI.73.12.7977-7987.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crisp RJ, Pollington A, Galea C, Jaron S, Yamaguchi-Iwai Y, Kaplan J. Inhibition of heme biosynthesis prevents transcription of iron uptake genes in yeast. J Biol Chem. 2003;278:45499–45506. doi: 10.1074/jbc.M307229200. [DOI] [PubMed] [Google Scholar]
- D'Souza CA, Alspaugh JA, Yue C, Harashima T, Cox GM, Perfect JR, Heitman J. Cyclic AMP-dependent protein kinase controls virulence of the fungal pathogen Cryptococcus neoformans. Mol Cell Biol. 2001;21:3179–3191. doi: 10.1128/MCB.21.9.3179-3191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson RC, Cruz MC, Sia RA, Allen B, Alspaugh JA, Heitman J. Gene disruption by biolistic transformation in serotype D strains of Cryptococcus neoformans. Fungal Genet Biol. 2000;29:38–48. doi: 10.1006/fgbi.1999.1180. [DOI] [PubMed] [Google Scholar]
- Davis DA. How human pathogenic fungi sense and adapt to pH: the link to virulence. Curr Opin Microbiol. 2009;12:365–70. doi: 10.1016/j.mib.2009.05.006. [DOI] [PubMed] [Google Scholar]
- Fang W, Price MS, Toffaletti DL, Tenor J, Betancourt-Quiroz M, Price JL, et al. Pleiotropic effects of deubiquitinating enzyme Ubp5 on growth and pathogenesis of Cryptococcus neoformans. PloS One. 2012;7:e38326. doi: 10.1371/journal.pone.0038326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster LA. Utilization and cell-surface binding of hemin by Histoplasma capsulatum. Can J Microbiol. 2002;48:437–442. doi: 10.1139/w02-037. [DOI] [PubMed] [Google Scholar]
- Godinho RM, Crestani J, Kmetzsch L, Araujo Gde S, Frases S, Staats CC, et al. The vacuolar-sorting protein Snf7 is required for export of virulence determinants in members of the Cryptococcus neoformans complex. Sci Rep. 2014;4:6198. doi: 10.1038/srep06198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths EJ, Hu G, Fries B, Caza M, Wang J, Gsponer J, et al. A defect in ATP-citrate lyase links acetyl-CoA production, virulence factor elaboration and virulence in Cryptococcus neoformans. Mol Microbiol. 2012;86:1404–1423. doi: 10.1111/mmi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han K, Do E, Jung WH. A human fungal pathogen Cryptococcus neoformans expresses three distinct iron permease homologs. J Microbiol Biotechnol. 2012;22:1644–1652. doi: 10.4014/jmb.1209.09019. [DOI] [PubMed] [Google Scholar]
- Hanson PI, Shim S, Merrill SA. Cell biology of the ESCRT machinery. Curr Op Cell Biol. 2009;21:568–574. doi: 10.1016/j.ceb.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Fukuzawa T, Sorimachi H, Maeda T. Constitutive activation of the pH-responsive Rim101 pathway in yeast mutants defective in late steps of the MVB/ESCRT pathway. Mol Cell Biol. 2005;25:9478–9490. doi: 10.1128/MCB.25.21.9478-9490.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henne WM, Buchkovich NJ, Emr SD. The ESCRT pathway. Dev Cell. 2011;21:77–91. doi: 10.1016/j.devcel.2011.05.015. [DOI] [PubMed] [Google Scholar]
- Hicks JK, D'Souza CA, Cox GM, Heitman J. Cyclic AMP-dependent protein kinase catalytic subunits have divergent roles in virulence factor production in two varieties of the fungal pathogen Cryptococcus neoformans. Eukaryot Cell. 2004;3:14–26. doi: 10.1128/EC.3.1.14-26.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu CJ, Bai C, Zheng XD, Wang YM, Wang Y. Characterization and functional analysis of the siderophore-iron transporter CaArn1p in Candida albicans. J Biol Chem. 2002;277:30598–30605. doi: 10.1074/jbc.M204545200. [DOI] [PubMed] [Google Scholar]
- Hu G, Caza M, Cadieux B, Chan V, Liu V, Kronstad J. Cryptococcus neoformans requires the ESCRT protein Vps23 for iron acquisition from heme, for capsule formation, and for virulence. Infect Immun. 2013;81:292–302. doi: 10.1128/IAI.01037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Cheng PY, Sham A, Perfect JR, Kronstad JW. Metabolic adaptation in Cryptococcus neoformans during early murine pulmonary infection. Mol Microbiol. 2008;69:1456–1475. doi: 10.1111/j.1365-2958.2008.06374.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Kronstad JW. A putative P-type ATPase, Apt1, is involved in stress tolerance and virulence in Cryptococcus neoformans. Eukaryot Cell. 2010;9:74–83. doi: 10.1128/EC.00289-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu G, Steen BR, Lian T, Sham AP, Tam N, Tangen KL, Kronstad JW. Transcriptional regulation by protein kinase A in Cryptococcus neoformans. PLoS Pathog. 2007;3:e42. doi: 10.1371/journal.ppat.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH. The ESCRT complexes. Crit Rev Biochem Mol Biol. 2010;45:463–487. doi: 10.3109/10409238.2010.502516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Emr SD. The ESCRT complexes: structure and mechanism of a membrane-trafficking network. Annu Rev Biophys Biomol Struct. 2006;35:277–298. doi: 10.1146/annurev.biophys.35.040405.102126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JH, Hanson PI. Membrane budding and scission by the ESCRT machinery: it's all in the neck. Nature Reviews Molecular Cell Biology. 2010;11:556–566. doi: 10.1038/nrm2937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito T, Chiba T, Ozawa R, Yoshida M, Hattori M, Sakaki Y. A comprehensive two-hybrid analysis to explore the yeast protein interactome. Proc Natl Acad Sci U S A. 2001;98:4569–4574. doi: 10.1073/pnas.061034498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Hu G, Kuo W, Kronstad JW. Role of ferroxidases in iron uptake and virulence of Cryptococcus neoformans. Eukaryot Cell. 2009;8:1511–1520. doi: 10.1128/EC.00166-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Kronstad JW. Iron and fungal pathogenesis: a case study with Cryptococcus neoformans. Cell Microbiol. 2008;10:277–284. doi: 10.1111/j.1462-5822.2007.01077.x. [DOI] [PubMed] [Google Scholar]
- Jung WH, Saikia S, Hu G, Wang J, Fung CK, D'Souza C, et al. HapX positively and negatively regulates the transcriptional response to iron deprivation in Cryptococcus neoformans. PLoS Pathog. 2010;6:e1001209. doi: 10.1371/journal.ppat.1001209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, Lian T, Singh A, Kosman DJ, Kronstad JW. Iron source preference and regulation of iron uptake in Cryptococcus neoformans. PLoS Pathog. 2008;4:e45. doi: 10.1371/journal.ppat.0040045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung WH, Sham A, White R, Kronstad JW. Iron regulation of the major virulence factors in the AIDS-associated pathogen Cryptococcus neoformans. PLoS Biol. 2006;4:e410. doi: 10.1371/journal.pbio.0040410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Cho YJ, Do E, Choi J, Hu G, Cadieux B, et al. A defect in iron uptake enhances the susceptibility of Cryptococcus neoformans to azole antifungal drugs. Fungal Genet Biol. 2012;49:955–966. doi: 10.1016/j.fgb.2012.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y, Yun CW, Philpott CC. Ferrichrome induces endosome to plasma membrane cycling of the ferrichrome transporter, Arn1p, in Saccharomyces cerevisiae. EMBO J. 2002;21:3632–3642. doi: 10.1093/emboj/cdf382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretschmer M, Reiner E, Hu G, Tam N, Oliveira DL, Caza M, et al. Defects in phosphate acquisition and storage influence virulence of Cryptococcus neoformans. Infect Immun. 2014;82:2697–2712. doi: 10.1128/IAI.01607-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad J, Saikia S, Nielson ED, Kretschmer M, Jung W, Hu G, et al. Adaptation of Cryptococcus neoformans to mammalian hosts: integrated regulation of metabolism and virulence. Eukaryot Cell. 2012;11:109–118. doi: 10.1128/EC.05273-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Attarian R, Cadieux B, Choi J, D'Souza CA, Griffiths EJ, et al. Expanding fungal pathogenesis: Cryptococcus breaks out of the opportunistic box. Nat Rev Microbiol. 2011a;9:193–203. doi: 10.1038/nrmicro2522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Hu G, Choi J. The cAMP/Protein Kinase A Pathway and Virulence in Cryptococcus neoformans. Mycobiology. 2011b;39:143–150. doi: 10.5941/MYCO.2011.39.3.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kronstad JW, Hu G, Jung WH. An encapsulation of iron homeostasis and virulence in Cryptococcus neoformans. Trends Microbiol. 2013;21:457–465. doi: 10.1016/j.tim.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuznets G, Vigonsky E, Weissman Z, Lalli D, Gildor T, Kauffman SJ, et al. A Relay Network of Extracellular Haem-Binding Proteins Drives C. albicans Iron Acquisition from Hemoglobin. PLoS Pathog. 2014;10:e1004407. doi: 10.1371/journal.ppat.1004407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lambert M, Blanchin-Roland S, Le Louedec F, Lepingle A, Gaillardin C. Genetic analysis of regulatory mutants affecting synthesis of extracellular proteinases in the yeast Yarrowia lipolytica: identification of a RIM101/pacC homolog. Mol Cell Biol. 1997;17:3966–3976. doi: 10.1128/mcb.17.7.3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W, Mitchell AP. Proteolytic activation of Rim1p, a positive regulator of yeast sporulation and invasive growth. Genetics. 1997;145:63–73. doi: 10.1093/genetics/145.1.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian T, Simmer MI, D'Souza CA, Steen BR, Zuyderduyn SD, Jones SJ, et al. Iron-regulated transcription and capsule formation in the fungal pathogen Cryptococcus neoformans. Mol Microbiol. 2005;55:1452–1472. doi: 10.1111/j.1365-2958.2004.04474.x. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001 2001 Dec;25(4):402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Loftus BJ, Fung E, Roncaglia P, Rowley D, Amedeo P, Bruno D, et al. The genome of the basidiomycetous yeast and human pathogen Cryptococcus neoformans. Science. 2005;307:1321–1324. doi: 10.1126/science.1103773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mingot JM, Tilburn J, Diez E, Bignell E, Orejas M, Widdick DA, et al. Specificity determinants of proteolytic processing of Aspergillus PacC transcription factor are remote from the processing site, and processing occurs in yeast if pH signalling is bypassed. Mol Cell Biol. 1999;19:1390–1400. doi: 10.1128/mcb.19.2.1390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita E, Sandrin V, McCullough J, Katsuyama A, Baci Hamilton I, Sundquist WI. ESCRT-III protein requirements for HIV-1 budding. Cell Host Microbe. 2011;9:235–242. doi: 10.1016/j.chom.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara TR, Alspaugh JA. The Cryptococcus neoformans capsule: a sword and a shield. Clin Microbiol Rev. 2012;25:387–408. doi: 10.1128/CMR.00001-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara TR, Holmer SM, Selvig K, Dietrich F, Alspaugh JA. Cryptococcus neoformans Rim101 is associated with cell wall remodeling and evasion of the host immune responses. MBio. 2013;4 doi: 10.1128/mBio.00522-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Meara TR, Norton D, Price MS, Hay C, Clements MF, Nichols CB, Alspaugh JA. Interaction of Cryptococcus neoformans Rim101 and protein kinase A regulates capsule. PLoS Pathog. 2010;6:e1000776. doi: 10.1371/journal.ppat.1000776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñalva MA, Tilburn J, Bignell E, Arst HN., Jr. Ambient pH gene regulation in fungi: making connections. Trends Microbiol. 2008;16:291–300. doi: 10.1016/j.tim.2008.03.006. [DOI] [PubMed] [Google Scholar]
- Pukkila-Worley R, Gerrald QD, Kraus PR, Boily MJ, Davis MJ, Giles SS, Cox GM, Heitman J, Alspaugh JA. Transcriptional network of multiple capsule and melanin genes governed by the Cryptococcus neoformans cyclic AMP cascade. Eukaryot Cell. 2005;4:190–201. doi: 10.1128/EC.4.1.190-201.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reese AJ, Doering TL. Cell wall alpha-1,3-glucan is required to anchor the Cryptococcus neoformans capsule. Mol Microbiol. 2003;50:1401–1409. doi: 10.1046/j.1365-2958.2003.03780.x. [DOI] [PubMed] [Google Scholar]
- Ren X, Hurley JH. VHS domains of ESCRT-0 cooperate in high-avidity binding to polyubiquitinated cargo. EMBO J. 2010;29:1045–1054. doi: 10.1038/emboj.2010.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia S, Oliveira D, Hu G, Kronstad JW. Role of ferric reductases in iron acquisition and virulence in the fungal pathogen Cryptococcus neoformans. Infect Immun. 2014;82:839–850. doi: 10.1128/IAI.01357-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos R, Buisson N, Knight S, Dancis A, Camadro JM, Lesuisse E. Haemin uptake and use as an iron source by Candida albicans: role of CaHMX1-encoded haem oxygenase. Microbiology. 2003;149:579–588. doi: 10.1099/mic.0.26108-0. [DOI] [PubMed] [Google Scholar]
- Stojiljkovic I, Kumar V, Srinivasan N. Non-iron metalloporphyrins: potent antibacterial compounds that exploit haem/Hb uptake systems of pathogenic bacteria. Mol Microbiol. 1999;31:429–442. doi: 10.1046/j.1365-2958.1999.01175.x. [DOI] [PubMed] [Google Scholar]
- Tangen KL, Jung WH, Sham AP, Lian T, Kronstad JW. The iron- and cAMP-regulated gene SIT1 influences ferrioxamine B utilization, melanization and cell wall structure in Cryptococcus neoformans. Microbiol. 2007;153:29–41. doi: 10.1099/mic.0.2006/000927-0. [DOI] [PubMed] [Google Scholar]
- Vincent O, Rainbow L, Tilburn J, Arst HN, Jr., Penalva MA. YPXL/I is a protein interaction motif recognized by Aspergillus PalA and its human homologue, AIP1/Alix. Mol Cell Biol. 2003;23:1647–1655. doi: 10.1128/MCB.23.5.1647-1655.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman Z, Kornitzer D. A family of Candida cell surface haem-binding proteins involved in haemin and haemoglobin-iron utilization. Mol Microbiol. 2004;53:1209–1220. doi: 10.1111/j.1365-2958.2004.04199.x. [DOI] [PubMed] [Google Scholar]
- Weissman Z, Shemer R, Conibear E, Kornitzer D. An endocytic mechanism for haemoglobin-iron acquisition in Candida albicans. Mol Microbiol. 2008;69:201–217. doi: 10.1111/j.1365-2958.2008.06277.x. [DOI] [PubMed] [Google Scholar]
- Wolf JM, Davis DA. Mutational analysis of Candida albicans SNF7 reveals genetically separable Rim101 and ESCRT functions and demonstrates divergence in bro1-domain protein interactions. Genetics. 2010;184:673–694. doi: 10.1534/genetics.109.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolf JM, Johnson DJ, Chmielewski D, Davis DA. The Candida albicans ESCRT pathway makes Rim101-dependent and -independent contributions to pathogenesis. Eukaryot Cell. 2010;9:1203–1215. doi: 10.1128/EC.00056-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wollert T, Wunder C, Lippincott-Schwartz J, Hurley JH. Membrane scission by the ESCRT-III complex. Nature. 2009;458:172–177. doi: 10.1038/nature07836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Mitchell AP. Yeast PalA/AIP1/Alix homolog Rim20p associates with a PEST-like region and is required for its proteolytic cleavage. J Bacteriol. 2001;183:6917–6923. doi: 10.1128/JB.183.23.6917-6923.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu W, Smith FJ, Jr., Subaran R, Mitchell AP. Multivesicular body-ESCRT components function in pH response regulation in Saccharomyces cerevisiae and Candida albicans. Mol Biol Cell. 2004;15:5528–5537. doi: 10.1091/mbc.E04-08-0666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoneda A, Doering TL. Regulation of Cryptococcus neoformans capsule size is mediated at the polymer level. Eukaryot Cell. 2008;7:546–549. doi: 10.1128/EC.00437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao Y, Du J, Xiong B, Xu H, Jiang L. ESCRT components regulate the expression of the ER/Golgi calcium pump gene PMR1 through the Rim101/Nrg1 pathway in budding yeast. J Mol Cell Biol. 2013;5:336–344. doi: 10.1093/jmcb/mjt025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure S1. Defective endocytosis and increased susceptibility to trafficking inhibitors for the ESCRT mutants. A. The WT and ESCRT mutant strains were grown in YPD medium at 30°C overnight, harvested, washed in PBS (pH 7.2), and stained with MDY-64 on ice for 15 min and washed to remove the unbound dye. The cells were then incubated for an additional 30 min at 30°C before viewing. The DIC (differential interference contrast) images are shown on the left and the fluorescence resulting from MDY64 staining is shown on the right. The results were also confirmed with FM4-64 staining (data not shown). B. Ten-fold serial dilutions of the indicated strains were spotted onto YNB alone or YNB supplemented with either 20 μg ml−1 or 40 μg ml−1 brefeldin A (BFA). The plates were incubated for 3 days at 30°C.
Supplemental Figure S2. Requirement of ESCRT components for the growth in liquid low iron medium supplemented with haem. Cells were inoculated into the liquid YNB + 150 μM BPS with and without ferric chloride or haem (10μM and 100μM), and then incubated at 30°C for 5 days. Optical density at 600 nm (OD600) were measured every 4 h using a NanoQuant Infinite M200 Pro TECAN system. Experiments were performed in triplicated. Each point stands for the expression of the mean of three measurements ± standard deviation (SD).
Supplementary Figure S3. Comparison of haem rescue of fluconazole susceptibility for mutants defective in ESCRT-I or high affinity iron uptake functions. Suppression of the fluconazole susceptibility of the vps23, cfo1, vps23 cfo1 and rim101 mutants was tested by addition of exogenous haem at 10 μM to YNB plus BPS medium containing 10 μg ml−1 fluconazole. For all assays, the strains were grown in low-iron medium and ten-fold serial dilutions of were spotted on agar plates, with subsequent incubation for 2-3 days at 30°C.
Supplementary Figure S4. Susceptibility of ESCRT mutants to agents that challenge cell wall integrity. Ten-fold serial dilutions of cells of the indicated strains were spotted onto YPD medium containing either 0.5 mg ml−1 caffeine, 0.5 mg ml−1 congo red, or 0.05% sodium dodecylsulfate (SDS). The plates were incubated for 3 days at 30°C.
Supplementary Figure S5. Reduced melanisation in ESCRT mutants. Ten-fold serial dilutions of cells of the indicated strains were spotted onto solid L-DOPA plates. The plates were incubated for 2 days at 30°C or 37°C prior to being photographed. Note, the ESCRT-III mutant, snf7, exhibits weak growth defect on YPD at 37°C, and has the most significant defect in melanisation. The vps4 and bro1 mutants produced a similar level of melanisation to the WT strain.
Supplemental Figure S6. Comparison of transcript levels for iron acquisition and haem uptake functions in response to iron deprivation. RNA levels for CIG1 were measured by quantitative RT-PCR analysis and compared for cells of the WT strain and the ESCRT mutants grown in low iron medium (YNB + BPS) versus iron-replete medium (YNB). The assays were performed twice in triplicate, and relative fold change values are reported as the mean ± SEM. The dashed lines indicate a two-fold change in transcript levels.